
Torque teno virus (isolate Human/China/CT39F/2001) (TTV)
Taxonomy: Viruses; Anelloviridae; unclassified Anelloviridae; Torque teno virus; TT virus group 5
Average proteome isoelectric point is 8.06
Get precalculated fractions of proteins
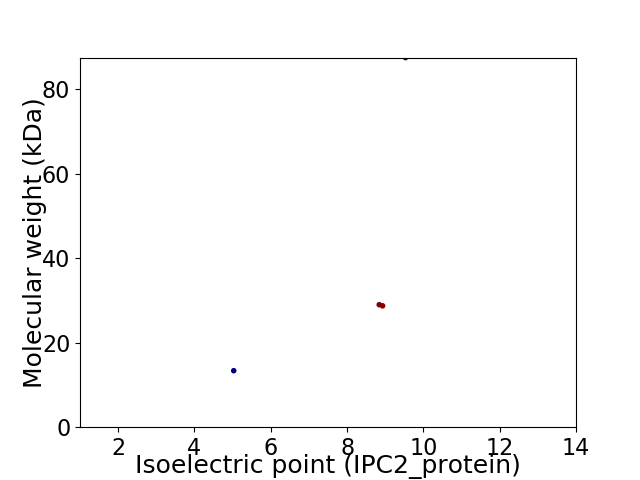
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q8V7G3|CAPSD_TTVV7 Capsid protein OS=Torque teno virus (isolate Human/China/CT39F/2001) OX=486279 GN=ORF1 PE=3 SV=1
MM1 pKa = 7.37SIWRR5 pKa = 11.84PPLHH9 pKa = 6.15NVPGLEE15 pKa = 4.08HH16 pKa = 6.59LWYY19 pKa = 9.8EE20 pKa = 4.45SVHH23 pKa = 6.19RR24 pKa = 11.84SHH26 pKa = 7.16AAVCGCGDD34 pKa = 3.83PVRR37 pKa = 11.84HH38 pKa = 5.3LTALAEE44 pKa = 4.4RR45 pKa = 11.84YY46 pKa = 8.75GIPGGSRR53 pKa = 11.84SSGAPGVGGNHH64 pKa = 6.24NPPQIRR70 pKa = 11.84RR71 pKa = 11.84ARR73 pKa = 11.84HH74 pKa = 5.02PAAAPDD80 pKa = 4.21PPAGNQPPALPWHH93 pKa = 6.68GDD95 pKa = 3.07GGNEE99 pKa = 3.89SGAGGGEE106 pKa = 4.08SGGPVADD113 pKa = 4.54FADD116 pKa = 5.0DD117 pKa = 4.2GLDD120 pKa = 3.75DD121 pKa = 5.35LVAALDD127 pKa = 3.81EE128 pKa = 4.67EE129 pKa = 4.8EE130 pKa = 4.1
MM1 pKa = 7.37SIWRR5 pKa = 11.84PPLHH9 pKa = 6.15NVPGLEE15 pKa = 4.08HH16 pKa = 6.59LWYY19 pKa = 9.8EE20 pKa = 4.45SVHH23 pKa = 6.19RR24 pKa = 11.84SHH26 pKa = 7.16AAVCGCGDD34 pKa = 3.83PVRR37 pKa = 11.84HH38 pKa = 5.3LTALAEE44 pKa = 4.4RR45 pKa = 11.84YY46 pKa = 8.75GIPGGSRR53 pKa = 11.84SSGAPGVGGNHH64 pKa = 6.24NPPQIRR70 pKa = 11.84RR71 pKa = 11.84ARR73 pKa = 11.84HH74 pKa = 5.02PAAAPDD80 pKa = 4.21PPAGNQPPALPWHH93 pKa = 6.68GDD95 pKa = 3.07GGNEE99 pKa = 3.89SGAGGGEE106 pKa = 4.08SGGPVADD113 pKa = 4.54FADD116 pKa = 5.0DD117 pKa = 4.2GLDD120 pKa = 3.75DD121 pKa = 5.35LVAALDD127 pKa = 3.81EE128 pKa = 4.67EE129 pKa = 4.8EE130 pKa = 4.1
Molecular weight: 13.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q8V7G4|ORF23_TTVV7 Probable protein VP2 OS=Torque teno virus (isolate Human/China/CT39F/2001) OX=486279 GN=ORF2/3 PE=3 SV=1
MM1 pKa = 7.53AWGWWKK7 pKa = 10.56RR8 pKa = 11.84KK9 pKa = 9.12RR10 pKa = 11.84RR11 pKa = 11.84WWWRR15 pKa = 11.84KK16 pKa = 6.64RR17 pKa = 11.84WTRR20 pKa = 11.84GRR22 pKa = 11.84LRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WPRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LYY56 pKa = 10.55RR57 pKa = 11.84RR58 pKa = 11.84GRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.06RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.81RR66 pKa = 11.84KK67 pKa = 8.89RR68 pKa = 11.84AKK70 pKa = 8.53ITIRR74 pKa = 11.84QWQPAMTRR82 pKa = 11.84RR83 pKa = 11.84CFIRR87 pKa = 11.84GHH89 pKa = 5.67MPALICGWGAYY100 pKa = 9.15ASNYY104 pKa = 7.97TSHH107 pKa = 8.24LEE109 pKa = 3.99DD110 pKa = 5.44KK111 pKa = 10.37IVKK114 pKa = 9.09GPYY117 pKa = 9.69GGGHH121 pKa = 4.82ATFRR125 pKa = 11.84FSLQVLCEE133 pKa = 4.03EE134 pKa = 4.64HH135 pKa = 6.8LKK137 pKa = 9.29HH138 pKa = 6.17HH139 pKa = 6.45NYY141 pKa = 6.52WTRR144 pKa = 11.84SNQDD148 pKa = 3.17LEE150 pKa = 4.06LALYY154 pKa = 10.05YY155 pKa = 10.81GATIKK160 pKa = 10.52FYY162 pKa = 10.74RR163 pKa = 11.84SPDD166 pKa = 2.79TDD168 pKa = 4.27FIVTYY173 pKa = 8.18QRR175 pKa = 11.84KK176 pKa = 9.02SPLGGNILTAPSLHH190 pKa = 6.78PAEE193 pKa = 5.48AMLSKK198 pKa = 10.91NKK200 pKa = 10.0ILIPSLQTKK209 pKa = 9.86PKK211 pKa = 9.63GKK213 pKa = 8.71KK214 pKa = 5.73TVKK217 pKa = 10.49VNIPPPTLFVHH228 pKa = 6.37KK229 pKa = 9.77WYY231 pKa = 9.46FQKK234 pKa = 10.69DD235 pKa = 3.31ICDD238 pKa = 3.36LTLFNLNVVAADD250 pKa = 3.82LRR252 pKa = 11.84FPFCSPQTDD261 pKa = 3.61NVCITFQVLAAEE273 pKa = 4.48YY274 pKa = 11.05NNFLSTTLGTTNEE287 pKa = 4.1STFIEE292 pKa = 4.42NFLKK296 pKa = 10.97VAFPDD301 pKa = 3.75DD302 pKa = 4.01KK303 pKa = 10.79PRR305 pKa = 11.84HH306 pKa = 5.42SNILNTFRR314 pKa = 11.84TEE316 pKa = 3.72GCMSHH321 pKa = 7.04PQLQKK326 pKa = 10.72FKK328 pKa = 10.44PPNTGPGEE336 pKa = 3.96NKK338 pKa = 10.33YY339 pKa = 10.81FFTPDD344 pKa = 3.97GLWGDD349 pKa = 4.53PIYY352 pKa = 10.43IYY354 pKa = 11.29NNGVQQQTAQQIRR367 pKa = 11.84EE368 pKa = 4.13KK369 pKa = 10.45IKK371 pKa = 10.85KK372 pKa = 9.99NMEE375 pKa = 3.52NYY377 pKa = 7.98YY378 pKa = 11.21AKK380 pKa = 10.05IVEE383 pKa = 4.56EE384 pKa = 3.91NTIITKK390 pKa = 10.18GSKK393 pKa = 9.16AHH395 pKa = 6.16CHH397 pKa = 4.42LTGIFSPPFLNIGRR411 pKa = 11.84VARR414 pKa = 11.84EE415 pKa = 3.74FPGLYY420 pKa = 9.52TDD422 pKa = 4.08VVYY425 pKa = 11.08NPWTDD430 pKa = 2.83KK431 pKa = 11.53GKK433 pKa = 11.09GNKK436 pKa = 8.59IWLDD440 pKa = 3.62SLTKK444 pKa = 10.23SDD446 pKa = 4.43NIYY449 pKa = 10.79DD450 pKa = 3.78PRR452 pKa = 11.84QSILLMADD460 pKa = 3.06MPLYY464 pKa = 10.66IMLNGYY470 pKa = 9.17IDD472 pKa = 3.43WAKK475 pKa = 10.79KK476 pKa = 9.0EE477 pKa = 4.07RR478 pKa = 11.84NNWGLATQYY487 pKa = 11.09RR488 pKa = 11.84LLLTCPYY495 pKa = 8.81TFPRR499 pKa = 11.84LYY501 pKa = 11.12VEE503 pKa = 4.25TNPNYY508 pKa = 10.67GYY510 pKa = 10.92VPYY513 pKa = 10.36SEE515 pKa = 5.27SFGAGQMPDD524 pKa = 3.23KK525 pKa = 10.85NPYY528 pKa = 9.6VPITWRR534 pKa = 11.84GKK536 pKa = 7.79WYY538 pKa = 9.28PHH540 pKa = 6.91ILHH543 pKa = 6.19QEE545 pKa = 3.8AVINDD550 pKa = 3.61IVISGPFTPKK560 pKa = 9.12DD561 pKa = 3.77TKK563 pKa = 10.84PVMQLNMKK571 pKa = 10.09YY572 pKa = 10.27SFRR575 pKa = 11.84FTWGGNPISTQIVKK589 pKa = 10.31DD590 pKa = 3.65PCTQPTFEE598 pKa = 4.86IPGGGNIPRR607 pKa = 11.84RR608 pKa = 11.84IQVINPKK615 pKa = 10.03VLGPSYY621 pKa = 10.85SFRR624 pKa = 11.84SFDD627 pKa = 3.44LRR629 pKa = 11.84RR630 pKa = 11.84DD631 pKa = 3.59MFSGSSLKK639 pKa = 10.31RR640 pKa = 11.84VSEE643 pKa = 3.99QQEE646 pKa = 3.92TSEE649 pKa = 4.4FLFSGGKK656 pKa = 9.25RR657 pKa = 11.84PRR659 pKa = 11.84IDD661 pKa = 3.2LPKK664 pKa = 10.0YY665 pKa = 9.87VPPEE669 pKa = 3.61EE670 pKa = 4.76DD671 pKa = 3.37FNIQEE676 pKa = 4.12RR677 pKa = 11.84QQRR680 pKa = 11.84EE681 pKa = 3.83QRR683 pKa = 11.84PWTSEE688 pKa = 4.03SEE690 pKa = 4.5SEE692 pKa = 4.79AEE694 pKa = 4.1AQEE697 pKa = 4.1EE698 pKa = 4.43TQAGSVRR705 pKa = 11.84EE706 pKa = 3.86QLQQQLQEE714 pKa = 4.06QFQLRR719 pKa = 11.84RR720 pKa = 11.84GLKK723 pKa = 10.11CLFEE727 pKa = 4.13QLVRR731 pKa = 11.84TQQGVHH737 pKa = 6.15VDD739 pKa = 3.41PCLVV743 pKa = 3.12
MM1 pKa = 7.53AWGWWKK7 pKa = 10.56RR8 pKa = 11.84KK9 pKa = 9.12RR10 pKa = 11.84RR11 pKa = 11.84WWWRR15 pKa = 11.84KK16 pKa = 6.64RR17 pKa = 11.84WTRR20 pKa = 11.84GRR22 pKa = 11.84LRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WPRR29 pKa = 11.84RR30 pKa = 11.84SRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84VRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84WRR47 pKa = 11.84RR48 pKa = 11.84GRR50 pKa = 11.84PRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84LYY56 pKa = 10.55RR57 pKa = 11.84RR58 pKa = 11.84GRR60 pKa = 11.84RR61 pKa = 11.84YY62 pKa = 9.06RR63 pKa = 11.84RR64 pKa = 11.84KK65 pKa = 9.81RR66 pKa = 11.84KK67 pKa = 8.89RR68 pKa = 11.84AKK70 pKa = 8.53ITIRR74 pKa = 11.84QWQPAMTRR82 pKa = 11.84RR83 pKa = 11.84CFIRR87 pKa = 11.84GHH89 pKa = 5.67MPALICGWGAYY100 pKa = 9.15ASNYY104 pKa = 7.97TSHH107 pKa = 8.24LEE109 pKa = 3.99DD110 pKa = 5.44KK111 pKa = 10.37IVKK114 pKa = 9.09GPYY117 pKa = 9.69GGGHH121 pKa = 4.82ATFRR125 pKa = 11.84FSLQVLCEE133 pKa = 4.03EE134 pKa = 4.64HH135 pKa = 6.8LKK137 pKa = 9.29HH138 pKa = 6.17HH139 pKa = 6.45NYY141 pKa = 6.52WTRR144 pKa = 11.84SNQDD148 pKa = 3.17LEE150 pKa = 4.06LALYY154 pKa = 10.05YY155 pKa = 10.81GATIKK160 pKa = 10.52FYY162 pKa = 10.74RR163 pKa = 11.84SPDD166 pKa = 2.79TDD168 pKa = 4.27FIVTYY173 pKa = 8.18QRR175 pKa = 11.84KK176 pKa = 9.02SPLGGNILTAPSLHH190 pKa = 6.78PAEE193 pKa = 5.48AMLSKK198 pKa = 10.91NKK200 pKa = 10.0ILIPSLQTKK209 pKa = 9.86PKK211 pKa = 9.63GKK213 pKa = 8.71KK214 pKa = 5.73TVKK217 pKa = 10.49VNIPPPTLFVHH228 pKa = 6.37KK229 pKa = 9.77WYY231 pKa = 9.46FQKK234 pKa = 10.69DD235 pKa = 3.31ICDD238 pKa = 3.36LTLFNLNVVAADD250 pKa = 3.82LRR252 pKa = 11.84FPFCSPQTDD261 pKa = 3.61NVCITFQVLAAEE273 pKa = 4.48YY274 pKa = 11.05NNFLSTTLGTTNEE287 pKa = 4.1STFIEE292 pKa = 4.42NFLKK296 pKa = 10.97VAFPDD301 pKa = 3.75DD302 pKa = 4.01KK303 pKa = 10.79PRR305 pKa = 11.84HH306 pKa = 5.42SNILNTFRR314 pKa = 11.84TEE316 pKa = 3.72GCMSHH321 pKa = 7.04PQLQKK326 pKa = 10.72FKK328 pKa = 10.44PPNTGPGEE336 pKa = 3.96NKK338 pKa = 10.33YY339 pKa = 10.81FFTPDD344 pKa = 3.97GLWGDD349 pKa = 4.53PIYY352 pKa = 10.43IYY354 pKa = 11.29NNGVQQQTAQQIRR367 pKa = 11.84EE368 pKa = 4.13KK369 pKa = 10.45IKK371 pKa = 10.85KK372 pKa = 9.99NMEE375 pKa = 3.52NYY377 pKa = 7.98YY378 pKa = 11.21AKK380 pKa = 10.05IVEE383 pKa = 4.56EE384 pKa = 3.91NTIITKK390 pKa = 10.18GSKK393 pKa = 9.16AHH395 pKa = 6.16CHH397 pKa = 4.42LTGIFSPPFLNIGRR411 pKa = 11.84VARR414 pKa = 11.84EE415 pKa = 3.74FPGLYY420 pKa = 9.52TDD422 pKa = 4.08VVYY425 pKa = 11.08NPWTDD430 pKa = 2.83KK431 pKa = 11.53GKK433 pKa = 11.09GNKK436 pKa = 8.59IWLDD440 pKa = 3.62SLTKK444 pKa = 10.23SDD446 pKa = 4.43NIYY449 pKa = 10.79DD450 pKa = 3.78PRR452 pKa = 11.84QSILLMADD460 pKa = 3.06MPLYY464 pKa = 10.66IMLNGYY470 pKa = 9.17IDD472 pKa = 3.43WAKK475 pKa = 10.79KK476 pKa = 9.0EE477 pKa = 4.07RR478 pKa = 11.84NNWGLATQYY487 pKa = 11.09RR488 pKa = 11.84LLLTCPYY495 pKa = 8.81TFPRR499 pKa = 11.84LYY501 pKa = 11.12VEE503 pKa = 4.25TNPNYY508 pKa = 10.67GYY510 pKa = 10.92VPYY513 pKa = 10.36SEE515 pKa = 5.27SFGAGQMPDD524 pKa = 3.23KK525 pKa = 10.85NPYY528 pKa = 9.6VPITWRR534 pKa = 11.84GKK536 pKa = 7.79WYY538 pKa = 9.28PHH540 pKa = 6.91ILHH543 pKa = 6.19QEE545 pKa = 3.8AVINDD550 pKa = 3.61IVISGPFTPKK560 pKa = 9.12DD561 pKa = 3.77TKK563 pKa = 10.84PVMQLNMKK571 pKa = 10.09YY572 pKa = 10.27SFRR575 pKa = 11.84FTWGGNPISTQIVKK589 pKa = 10.31DD590 pKa = 3.65PCTQPTFEE598 pKa = 4.86IPGGGNIPRR607 pKa = 11.84RR608 pKa = 11.84IQVINPKK615 pKa = 10.03VLGPSYY621 pKa = 10.85SFRR624 pKa = 11.84SFDD627 pKa = 3.44LRR629 pKa = 11.84RR630 pKa = 11.84DD631 pKa = 3.59MFSGSSLKK639 pKa = 10.31RR640 pKa = 11.84VSEE643 pKa = 3.99QQEE646 pKa = 3.92TSEE649 pKa = 4.4FLFSGGKK656 pKa = 9.25RR657 pKa = 11.84PRR659 pKa = 11.84IDD661 pKa = 3.2LPKK664 pKa = 10.0YY665 pKa = 9.87VPPEE669 pKa = 3.61EE670 pKa = 4.76DD671 pKa = 3.37FNIQEE676 pKa = 4.12RR677 pKa = 11.84QQRR680 pKa = 11.84EE681 pKa = 3.83QRR683 pKa = 11.84PWTSEE688 pKa = 4.03SEE690 pKa = 4.5SEE692 pKa = 4.79AEE694 pKa = 4.1AQEE697 pKa = 4.1EE698 pKa = 4.43TQAGSVRR705 pKa = 11.84EE706 pKa = 3.86QLQQQLQEE714 pKa = 4.06QFQLRR719 pKa = 11.84RR720 pKa = 11.84GLKK723 pKa = 10.11CLFEE727 pKa = 4.13QLVRR731 pKa = 11.84TQQGVHH737 pKa = 6.15VDD739 pKa = 3.41PCLVV743 pKa = 3.12
Molecular weight: 87.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1411 |
130 |
743 |
352.8 |
39.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.229 ± 2.238 | 1.488 ± 0.099 |
4.536 ± 0.606 | 5.174 ± 0.251 |
3.118 ± 1.138 | 8.434 ± 1.729 |
2.764 ± 0.689 | 3.827 ± 1.174 |
5.103 ± 1.14 | 7.158 ± 0.244 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.347 ± 0.328 | 4.607 ± 0.34 |
9.497 ± 1.439 | 4.04 ± 1.109 |
9.284 ± 0.752 | 7.3 ± 1.895 |
4.748 ± 1.164 | 4.607 ± 0.157 |
2.339 ± 0.412 | 3.402 ± 0.748 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
