
Prolixibacteraceae bacterium WC007
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Prolixibacteraceae; Maribellus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
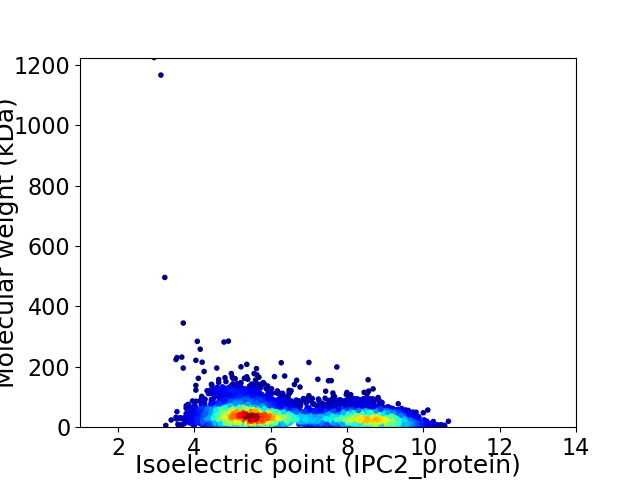
Virtual 2D-PAGE plot for 6116 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6I6JYM3|A0A6I6JYM3_9BACT Xylose isomerase OS=Prolixibacteraceae bacterium WC007 OX=2681766 GN=xylA PE=3 SV=1
MM1 pKa = 7.5RR2 pKa = 11.84VKK4 pKa = 10.18TEE6 pKa = 3.84TFKK9 pKa = 11.51NMKK12 pKa = 9.47KK13 pKa = 9.05WFFYY17 pKa = 9.27AWAVVTVLFASCEE30 pKa = 3.98GLTPGTGTEE39 pKa = 4.18EE40 pKa = 4.55EE41 pKa = 4.26EE42 pKa = 4.22QQDD45 pKa = 3.94VVNEE49 pKa = 4.07EE50 pKa = 4.35TIEE53 pKa = 3.98EE54 pKa = 4.28AVTNNAGDD62 pKa = 4.08HH63 pKa = 7.52DD64 pKa = 4.44DD65 pKa = 3.88TADD68 pKa = 3.69YY69 pKa = 9.22TWDD72 pKa = 3.89SNSVTQISLNGSSATEE88 pKa = 3.94SSDD91 pKa = 3.94DD92 pKa = 3.61VTIEE96 pKa = 3.89SGNIVIGAAGNYY108 pKa = 9.23SFSGTLSNGQIIVNTEE124 pKa = 3.53DD125 pKa = 3.46EE126 pKa = 4.72EE127 pKa = 4.42IVRR130 pKa = 11.84LILDD134 pKa = 4.2GVDD137 pKa = 3.8ISCSNSAPIYY147 pKa = 9.67IKK149 pKa = 10.59SAEE152 pKa = 4.17KK153 pKa = 10.83VLIALNEE160 pKa = 3.95NTTNKK165 pKa = 8.89LTDD168 pKa = 3.86GSSYY172 pKa = 11.36NYY174 pKa = 10.24DD175 pKa = 3.53DD176 pKa = 5.71EE177 pKa = 5.52EE178 pKa = 5.86DD179 pKa = 4.07EE180 pKa = 4.74EE181 pKa = 4.51PNAAIFSKK189 pKa = 10.95SDD191 pKa = 3.12LTIFGEE197 pKa = 4.54GTLVVDD203 pKa = 4.92ANFNDD208 pKa = 5.64GITSKK213 pKa = 10.89DD214 pKa = 3.32GLIIASGNISVTAVDD229 pKa = 3.99DD230 pKa = 4.91GIRR233 pKa = 11.84GKK235 pKa = 10.58DD236 pKa = 3.33YY237 pKa = 10.96IIIKK241 pKa = 10.05NGNFTVEE248 pKa = 3.92SDD250 pKa = 3.47GDD252 pKa = 4.07GLKK255 pKa = 10.39SDD257 pKa = 4.64NDD259 pKa = 3.79SDD261 pKa = 4.1DD262 pKa = 3.57SKK264 pKa = 11.75GYY266 pKa = 10.26IEE268 pKa = 5.5IYY270 pKa = 10.58DD271 pKa = 3.66GTFDD275 pKa = 4.48ISAKK279 pKa = 9.97GDD281 pKa = 3.51AVSAEE286 pKa = 4.0TDD288 pKa = 3.38LMIEE292 pKa = 4.03YY293 pKa = 10.86AEE295 pKa = 4.55IEE297 pKa = 4.37LTTSGSTSSYY307 pKa = 11.01YY308 pKa = 10.68EE309 pKa = 4.04STSSKK314 pKa = 10.46GLKK317 pKa = 10.12AGVNIIIDD325 pKa = 3.83DD326 pKa = 4.9GIFSLNCADD335 pKa = 5.66DD336 pKa = 5.73AIHH339 pKa = 6.78SNQTITINSGTYY351 pKa = 9.46EE352 pKa = 4.11ISSGDD357 pKa = 3.76DD358 pKa = 4.38GIHH361 pKa = 6.61SDD363 pKa = 3.94YY364 pKa = 11.66DD365 pKa = 3.68LVINDD370 pKa = 3.76GDD372 pKa = 3.89INITKK377 pKa = 9.62SYY379 pKa = 10.57EE380 pKa = 4.36GIEE383 pKa = 4.15SGQGDD388 pKa = 3.84MEE390 pKa = 4.76INKK393 pKa = 8.39GTIHH397 pKa = 6.69IKK399 pKa = 10.73SSDD402 pKa = 3.44DD403 pKa = 4.38GINLSAGGDD412 pKa = 3.47AMGGGGPWGGGSTSSGNYY430 pKa = 9.29YY431 pKa = 9.99IYY433 pKa = 10.63INGAYY438 pKa = 9.28IYY440 pKa = 10.13INASGDD446 pKa = 3.76GLDD449 pKa = 3.56SNGNIKK455 pKa = 8.02MTDD458 pKa = 3.02GTVLVDD464 pKa = 4.14GPTNSGNGALDD475 pKa = 3.81CNGSFTITGGLLIAAGSSGMAEE497 pKa = 4.31APASSSDD504 pKa = 3.57QYY506 pKa = 11.88SVLVKK511 pKa = 10.39FNSTKK516 pKa = 10.05SGGTIFHH523 pKa = 6.28VEE525 pKa = 4.03SEE527 pKa = 4.14KK528 pKa = 11.32GEE530 pKa = 4.31EE531 pKa = 3.75ILTYY535 pKa = 10.27KK536 pKa = 10.25PSKK539 pKa = 10.13NYY541 pKa = 10.22QSVAFSSPDD550 pKa = 3.25LTKK553 pKa = 10.93GEE555 pKa = 4.53SYY557 pKa = 10.77SIYY560 pKa = 10.21TGGSSTGDD568 pKa = 3.25EE569 pKa = 4.29TDD571 pKa = 3.47GLFSGGTYY579 pKa = 9.87TPGSVYY585 pKa = 10.64VSFDD589 pKa = 3.4ITSMTYY595 pKa = 9.91TIQQ598 pKa = 3.44
MM1 pKa = 7.5RR2 pKa = 11.84VKK4 pKa = 10.18TEE6 pKa = 3.84TFKK9 pKa = 11.51NMKK12 pKa = 9.47KK13 pKa = 9.05WFFYY17 pKa = 9.27AWAVVTVLFASCEE30 pKa = 3.98GLTPGTGTEE39 pKa = 4.18EE40 pKa = 4.55EE41 pKa = 4.26EE42 pKa = 4.22QQDD45 pKa = 3.94VVNEE49 pKa = 4.07EE50 pKa = 4.35TIEE53 pKa = 3.98EE54 pKa = 4.28AVTNNAGDD62 pKa = 4.08HH63 pKa = 7.52DD64 pKa = 4.44DD65 pKa = 3.88TADD68 pKa = 3.69YY69 pKa = 9.22TWDD72 pKa = 3.89SNSVTQISLNGSSATEE88 pKa = 3.94SSDD91 pKa = 3.94DD92 pKa = 3.61VTIEE96 pKa = 3.89SGNIVIGAAGNYY108 pKa = 9.23SFSGTLSNGQIIVNTEE124 pKa = 3.53DD125 pKa = 3.46EE126 pKa = 4.72EE127 pKa = 4.42IVRR130 pKa = 11.84LILDD134 pKa = 4.2GVDD137 pKa = 3.8ISCSNSAPIYY147 pKa = 9.67IKK149 pKa = 10.59SAEE152 pKa = 4.17KK153 pKa = 10.83VLIALNEE160 pKa = 3.95NTTNKK165 pKa = 8.89LTDD168 pKa = 3.86GSSYY172 pKa = 11.36NYY174 pKa = 10.24DD175 pKa = 3.53DD176 pKa = 5.71EE177 pKa = 5.52EE178 pKa = 5.86DD179 pKa = 4.07EE180 pKa = 4.74EE181 pKa = 4.51PNAAIFSKK189 pKa = 10.95SDD191 pKa = 3.12LTIFGEE197 pKa = 4.54GTLVVDD203 pKa = 4.92ANFNDD208 pKa = 5.64GITSKK213 pKa = 10.89DD214 pKa = 3.32GLIIASGNISVTAVDD229 pKa = 3.99DD230 pKa = 4.91GIRR233 pKa = 11.84GKK235 pKa = 10.58DD236 pKa = 3.33YY237 pKa = 10.96IIIKK241 pKa = 10.05NGNFTVEE248 pKa = 3.92SDD250 pKa = 3.47GDD252 pKa = 4.07GLKK255 pKa = 10.39SDD257 pKa = 4.64NDD259 pKa = 3.79SDD261 pKa = 4.1DD262 pKa = 3.57SKK264 pKa = 11.75GYY266 pKa = 10.26IEE268 pKa = 5.5IYY270 pKa = 10.58DD271 pKa = 3.66GTFDD275 pKa = 4.48ISAKK279 pKa = 9.97GDD281 pKa = 3.51AVSAEE286 pKa = 4.0TDD288 pKa = 3.38LMIEE292 pKa = 4.03YY293 pKa = 10.86AEE295 pKa = 4.55IEE297 pKa = 4.37LTTSGSTSSYY307 pKa = 11.01YY308 pKa = 10.68EE309 pKa = 4.04STSSKK314 pKa = 10.46GLKK317 pKa = 10.12AGVNIIIDD325 pKa = 3.83DD326 pKa = 4.9GIFSLNCADD335 pKa = 5.66DD336 pKa = 5.73AIHH339 pKa = 6.78SNQTITINSGTYY351 pKa = 9.46EE352 pKa = 4.11ISSGDD357 pKa = 3.76DD358 pKa = 4.38GIHH361 pKa = 6.61SDD363 pKa = 3.94YY364 pKa = 11.66DD365 pKa = 3.68LVINDD370 pKa = 3.76GDD372 pKa = 3.89INITKK377 pKa = 9.62SYY379 pKa = 10.57EE380 pKa = 4.36GIEE383 pKa = 4.15SGQGDD388 pKa = 3.84MEE390 pKa = 4.76INKK393 pKa = 8.39GTIHH397 pKa = 6.69IKK399 pKa = 10.73SSDD402 pKa = 3.44DD403 pKa = 4.38GINLSAGGDD412 pKa = 3.47AMGGGGPWGGGSTSSGNYY430 pKa = 9.29YY431 pKa = 9.99IYY433 pKa = 10.63INGAYY438 pKa = 9.28IYY440 pKa = 10.13INASGDD446 pKa = 3.76GLDD449 pKa = 3.56SNGNIKK455 pKa = 8.02MTDD458 pKa = 3.02GTVLVDD464 pKa = 4.14GPTNSGNGALDD475 pKa = 3.81CNGSFTITGGLLIAAGSSGMAEE497 pKa = 4.31APASSSDD504 pKa = 3.57QYY506 pKa = 11.88SVLVKK511 pKa = 10.39FNSTKK516 pKa = 10.05SGGTIFHH523 pKa = 6.28VEE525 pKa = 4.03SEE527 pKa = 4.14KK528 pKa = 11.32GEE530 pKa = 4.31EE531 pKa = 3.75ILTYY535 pKa = 10.27KK536 pKa = 10.25PSKK539 pKa = 10.13NYY541 pKa = 10.22QSVAFSSPDD550 pKa = 3.25LTKK553 pKa = 10.93GEE555 pKa = 4.53SYY557 pKa = 10.77SIYY560 pKa = 10.21TGGSSTGDD568 pKa = 3.25EE569 pKa = 4.29TDD571 pKa = 3.47GLFSGGTYY579 pKa = 9.87TPGSVYY585 pKa = 10.64VSFDD589 pKa = 3.4ITSMTYY595 pKa = 9.91TIQQ598 pKa = 3.44
Molecular weight: 62.96 kDa
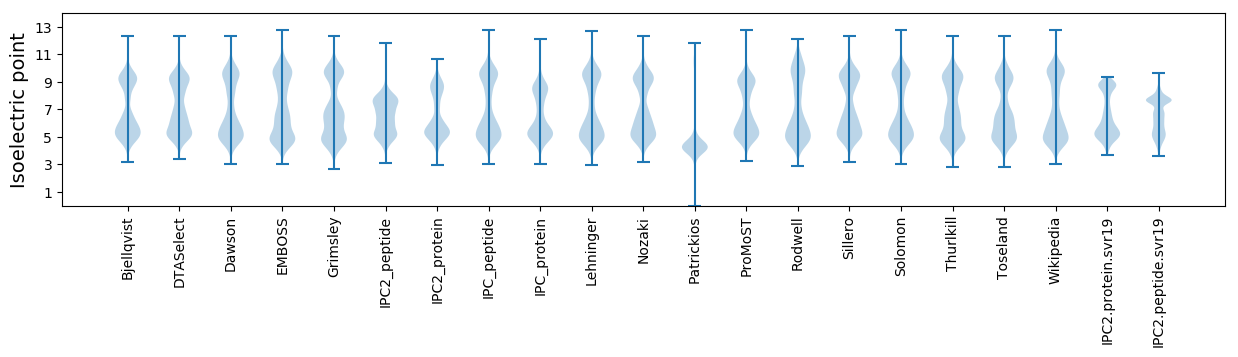
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6I6JWB1|A0A6I6JWB1_9BACT Mandelate racemase/muconate lactonizing enzyme family protein OS=Prolixibacteraceae bacterium WC007 OX=2681766 GN=GM418_18555 PE=4 SV=1
MM1 pKa = 7.93AKK3 pKa = 9.78RR4 pKa = 11.84ISIIIINLGIVVWGIIGFNHH24 pKa = 5.99LNYY27 pKa = 9.36WEE29 pKa = 4.17RR30 pKa = 11.84SARR33 pKa = 11.84IFQVNSEE40 pKa = 4.19QNFRR44 pKa = 11.84RR45 pKa = 11.84GFDD48 pKa = 2.86RR49 pKa = 11.84GRR51 pKa = 11.84PGFEE55 pKa = 5.24DD56 pKa = 3.66RR57 pKa = 11.84SSWDD61 pKa = 2.95RR62 pKa = 11.84STLTEE67 pKa = 5.24RR68 pKa = 11.84PDD70 pKa = 3.76LLNLSDD76 pKa = 6.04SIAQQSADD84 pKa = 3.11VVQRR88 pKa = 11.84NRR90 pKa = 11.84EE91 pKa = 4.24SIRR94 pKa = 11.84EE95 pKa = 4.02RR96 pKa = 11.84NSEE99 pKa = 3.7DD100 pKa = 2.85RR101 pKa = 11.84SYY103 pKa = 11.55GRR105 pKa = 11.84RR106 pKa = 11.84GFHH109 pKa = 6.4SRR111 pKa = 11.84QKK113 pKa = 8.86IQLRR117 pKa = 11.84NVVWFLAVFCLFTVGTIYY135 pKa = 10.44TDD137 pKa = 3.02KK138 pKa = 10.52TIRR141 pKa = 11.84AFRR144 pKa = 11.84KK145 pKa = 9.53KK146 pKa = 10.16EE147 pKa = 3.76KK148 pKa = 10.67
MM1 pKa = 7.93AKK3 pKa = 9.78RR4 pKa = 11.84ISIIIINLGIVVWGIIGFNHH24 pKa = 5.99LNYY27 pKa = 9.36WEE29 pKa = 4.17RR30 pKa = 11.84SARR33 pKa = 11.84IFQVNSEE40 pKa = 4.19QNFRR44 pKa = 11.84RR45 pKa = 11.84GFDD48 pKa = 2.86RR49 pKa = 11.84GRR51 pKa = 11.84PGFEE55 pKa = 5.24DD56 pKa = 3.66RR57 pKa = 11.84SSWDD61 pKa = 2.95RR62 pKa = 11.84STLTEE67 pKa = 5.24RR68 pKa = 11.84PDD70 pKa = 3.76LLNLSDD76 pKa = 6.04SIAQQSADD84 pKa = 3.11VVQRR88 pKa = 11.84NRR90 pKa = 11.84EE91 pKa = 4.24SIRR94 pKa = 11.84EE95 pKa = 4.02RR96 pKa = 11.84NSEE99 pKa = 3.7DD100 pKa = 2.85RR101 pKa = 11.84SYY103 pKa = 11.55GRR105 pKa = 11.84RR106 pKa = 11.84GFHH109 pKa = 6.4SRR111 pKa = 11.84QKK113 pKa = 8.86IQLRR117 pKa = 11.84NVVWFLAVFCLFTVGTIYY135 pKa = 10.44TDD137 pKa = 3.02KK138 pKa = 10.52TIRR141 pKa = 11.84AFRR144 pKa = 11.84KK145 pKa = 9.53KK146 pKa = 10.16EE147 pKa = 3.76KK148 pKa = 10.67
Molecular weight: 17.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2299017 |
32 |
11651 |
375.9 |
42.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.113 ± 0.046 | 0.855 ± 0.028 |
5.495 ± 0.052 | 7.0 ± 0.039 |
5.344 ± 0.028 | 6.726 ± 0.033 |
1.702 ± 0.015 | 7.582 ± 0.03 |
7.387 ± 0.054 | 8.949 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.018 | 5.985 ± 0.028 |
3.742 ± 0.02 | 3.35 ± 0.017 |
3.772 ± 0.028 | 6.488 ± 0.025 |
5.472 ± 0.073 | 6.28 ± 0.022 |
1.384 ± 0.014 | 4.144 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
