
Pacific flying fox faeces associated circular DNA virus-10
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.7
Get precalculated fractions of proteins
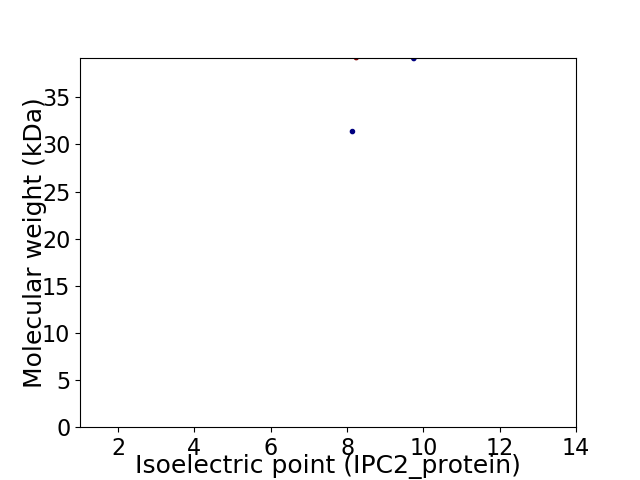
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
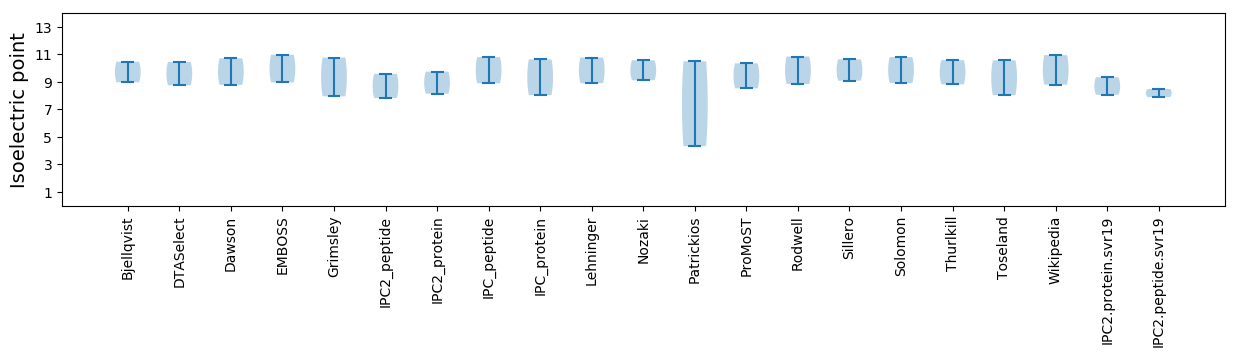
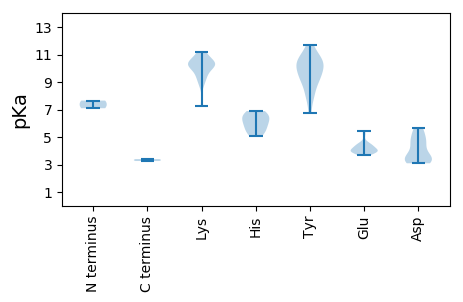
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTU9|A0A140CTU9_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-10 OX=1796005 PE=3 SV=1
MM1 pKa = 7.1SRR3 pKa = 11.84VRR5 pKa = 11.84VWVWTSFKK13 pKa = 11.07DD14 pKa = 4.68SLDD17 pKa = 4.01LNDD20 pKa = 4.73GGIEE24 pKa = 3.93YY25 pKa = 10.35SVYY28 pKa = 10.12QQEE31 pKa = 4.28QCPTTGRR38 pKa = 11.84LHH40 pKa = 5.45WQGVTRR46 pKa = 11.84FKK48 pKa = 10.88NARR51 pKa = 11.84QLGGVKK57 pKa = 9.8RR58 pKa = 11.84WLGDD62 pKa = 3.52PQAHH66 pKa = 6.68CEE68 pKa = 4.08PCKK71 pKa = 10.41KK72 pKa = 9.41EE73 pKa = 3.87HH74 pKa = 6.35EE75 pKa = 4.62AIKK78 pKa = 10.35YY79 pKa = 6.73CQKK82 pKa = 10.31EE83 pKa = 4.01EE84 pKa = 4.06TRR86 pKa = 11.84IGQATILGHH95 pKa = 5.04IQEE98 pKa = 4.52TLEE101 pKa = 4.3PGWWQKK107 pKa = 9.12LTINQLWEE115 pKa = 3.91QHH117 pKa = 5.98PEE119 pKa = 3.68WMLKK123 pKa = 10.08HH124 pKa = 5.46YY125 pKa = 10.87NGVQAYY131 pKa = 8.95RR132 pKa = 11.84RR133 pKa = 11.84SLNKK137 pKa = 10.36SQTEE141 pKa = 3.91RR142 pKa = 11.84QNIKK146 pKa = 10.14VFVFYY151 pKa = 8.01GTPGTGKK158 pKa = 10.16SYY160 pKa = 10.67SARR163 pKa = 11.84QFKK166 pKa = 10.36PYY168 pKa = 9.26FAKK171 pKa = 10.83ASGNWWDD178 pKa = 4.72GYY180 pKa = 10.7CGEE183 pKa = 4.43EE184 pKa = 3.93TVIFDD189 pKa = 4.48DD190 pKa = 5.64FYY192 pKa = 11.68GANEE196 pKa = 4.5KK197 pKa = 9.33YY198 pKa = 9.41TDD200 pKa = 3.26ILRR203 pKa = 11.84WCSEE207 pKa = 4.04LPISVPIKK215 pKa = 10.35GSMVPLEE222 pKa = 4.09ATTFIFTSNSHH233 pKa = 6.89PSLWWPGLLDD243 pKa = 3.21KK244 pKa = 11.17SAFQRR249 pKa = 11.84RR250 pKa = 11.84VTRR253 pKa = 11.84IYY255 pKa = 10.18QCFIEE260 pKa = 5.14KK261 pKa = 9.74FVKK264 pKa = 10.15QVWW267 pKa = 3.26
MM1 pKa = 7.1SRR3 pKa = 11.84VRR5 pKa = 11.84VWVWTSFKK13 pKa = 11.07DD14 pKa = 4.68SLDD17 pKa = 4.01LNDD20 pKa = 4.73GGIEE24 pKa = 3.93YY25 pKa = 10.35SVYY28 pKa = 10.12QQEE31 pKa = 4.28QCPTTGRR38 pKa = 11.84LHH40 pKa = 5.45WQGVTRR46 pKa = 11.84FKK48 pKa = 10.88NARR51 pKa = 11.84QLGGVKK57 pKa = 9.8RR58 pKa = 11.84WLGDD62 pKa = 3.52PQAHH66 pKa = 6.68CEE68 pKa = 4.08PCKK71 pKa = 10.41KK72 pKa = 9.41EE73 pKa = 3.87HH74 pKa = 6.35EE75 pKa = 4.62AIKK78 pKa = 10.35YY79 pKa = 6.73CQKK82 pKa = 10.31EE83 pKa = 4.01EE84 pKa = 4.06TRR86 pKa = 11.84IGQATILGHH95 pKa = 5.04IQEE98 pKa = 4.52TLEE101 pKa = 4.3PGWWQKK107 pKa = 9.12LTINQLWEE115 pKa = 3.91QHH117 pKa = 5.98PEE119 pKa = 3.68WMLKK123 pKa = 10.08HH124 pKa = 5.46YY125 pKa = 10.87NGVQAYY131 pKa = 8.95RR132 pKa = 11.84RR133 pKa = 11.84SLNKK137 pKa = 10.36SQTEE141 pKa = 3.91RR142 pKa = 11.84QNIKK146 pKa = 10.14VFVFYY151 pKa = 8.01GTPGTGKK158 pKa = 10.16SYY160 pKa = 10.67SARR163 pKa = 11.84QFKK166 pKa = 10.36PYY168 pKa = 9.26FAKK171 pKa = 10.83ASGNWWDD178 pKa = 4.72GYY180 pKa = 10.7CGEE183 pKa = 4.43EE184 pKa = 3.93TVIFDD189 pKa = 4.48DD190 pKa = 5.64FYY192 pKa = 11.68GANEE196 pKa = 4.5KK197 pKa = 9.33YY198 pKa = 9.41TDD200 pKa = 3.26ILRR203 pKa = 11.84WCSEE207 pKa = 4.04LPISVPIKK215 pKa = 10.35GSMVPLEE222 pKa = 4.09ATTFIFTSNSHH233 pKa = 6.89PSLWWPGLLDD243 pKa = 3.21KK244 pKa = 11.17SAFQRR249 pKa = 11.84RR250 pKa = 11.84VTRR253 pKa = 11.84IYY255 pKa = 10.18QCFIEE260 pKa = 5.14KK261 pKa = 9.74FVKK264 pKa = 10.15QVWW267 pKa = 3.26
Molecular weight: 31.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTU9|A0A140CTU9_9VIRU ATP-dependent helicase Rep OS=Pacific flying fox faeces associated circular DNA virus-10 OX=1796005 PE=3 SV=1
MM1 pKa = 7.65AIVKK5 pKa = 9.56RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VIRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PVIRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 7.28YY22 pKa = 9.1GRR24 pKa = 11.84VARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84LGRR32 pKa = 11.84RR33 pKa = 11.84SVRR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FAALPNWSKK52 pKa = 11.14RR53 pKa = 11.84DD54 pKa = 3.42YY55 pKa = 11.41SGISNLSNYY64 pKa = 9.43YY65 pKa = 10.15VVAQEE70 pKa = 4.3TPTVEE75 pKa = 3.92NSWYY79 pKa = 10.59HH80 pKa = 6.26LVFTQAQLYY89 pKa = 8.07PAFVQLWKK97 pKa = 10.35EE98 pKa = 3.69YY99 pKa = 8.92RR100 pKa = 11.84LAYY103 pKa = 10.1AVIAVRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84NPQAVVYY118 pKa = 7.97STSSGSVTASRR129 pKa = 11.84VEE131 pKa = 4.45SNNIWWAPWAKK142 pKa = 10.15YY143 pKa = 9.72DD144 pKa = 5.2PPVQPPRR151 pKa = 11.84MVRR154 pKa = 11.84SAVLLTEE161 pKa = 4.18RR162 pKa = 11.84WTVRR166 pKa = 11.84KK167 pKa = 8.78LANKK171 pKa = 9.45CPYY174 pKa = 10.43YY175 pKa = 10.65DD176 pKa = 5.62FSWQPPFGTSTDD188 pKa = 3.11ISGVPDD194 pKa = 3.4IKK196 pKa = 10.8IPSRR200 pKa = 11.84VIAASLFDD208 pKa = 5.39ADD210 pKa = 4.04TTTPTSSEE218 pKa = 3.76QSLLKK223 pKa = 10.3TFGQFTARR231 pKa = 11.84WRR233 pKa = 11.84SVPFQSVDD241 pKa = 3.18TQEE244 pKa = 4.37QVVHH248 pKa = 6.61CGFFCTEE255 pKa = 4.02NKK257 pKa = 10.3SAIQPDD263 pKa = 4.73LIEE266 pKa = 3.96VCARR270 pKa = 11.84VVFQMRR276 pKa = 11.84GRR278 pKa = 11.84KK279 pKa = 8.84NLGTEE284 pKa = 4.23TDD286 pKa = 3.09SSGAFDD292 pKa = 3.34AAEE295 pKa = 3.92VVEE298 pKa = 5.4FSMDD302 pKa = 3.37QRR304 pKa = 11.84GTAPPKK310 pKa = 10.32LRR312 pKa = 11.84YY313 pKa = 9.62GPLKK317 pKa = 10.46ALPDD321 pKa = 3.74CSSFKK326 pKa = 10.46TSNPTDD332 pKa = 3.38LHH334 pKa = 5.86VQQITCSS341 pKa = 3.4
MM1 pKa = 7.65AIVKK5 pKa = 9.56RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84VIRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84PVIRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 7.28YY22 pKa = 9.1GRR24 pKa = 11.84VARR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84LGRR32 pKa = 11.84RR33 pKa = 11.84SVRR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FAALPNWSKK52 pKa = 11.14RR53 pKa = 11.84DD54 pKa = 3.42YY55 pKa = 11.41SGISNLSNYY64 pKa = 9.43YY65 pKa = 10.15VVAQEE70 pKa = 4.3TPTVEE75 pKa = 3.92NSWYY79 pKa = 10.59HH80 pKa = 6.26LVFTQAQLYY89 pKa = 8.07PAFVQLWKK97 pKa = 10.35EE98 pKa = 3.69YY99 pKa = 8.92RR100 pKa = 11.84LAYY103 pKa = 10.1AVIAVRR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84NPQAVVYY118 pKa = 7.97STSSGSVTASRR129 pKa = 11.84VEE131 pKa = 4.45SNNIWWAPWAKK142 pKa = 10.15YY143 pKa = 9.72DD144 pKa = 5.2PPVQPPRR151 pKa = 11.84MVRR154 pKa = 11.84SAVLLTEE161 pKa = 4.18RR162 pKa = 11.84WTVRR166 pKa = 11.84KK167 pKa = 8.78LANKK171 pKa = 9.45CPYY174 pKa = 10.43YY175 pKa = 10.65DD176 pKa = 5.62FSWQPPFGTSTDD188 pKa = 3.11ISGVPDD194 pKa = 3.4IKK196 pKa = 10.8IPSRR200 pKa = 11.84VIAASLFDD208 pKa = 5.39ADD210 pKa = 4.04TTTPTSSEE218 pKa = 3.76QSLLKK223 pKa = 10.3TFGQFTARR231 pKa = 11.84WRR233 pKa = 11.84SVPFQSVDD241 pKa = 3.18TQEE244 pKa = 4.37QVVHH248 pKa = 6.61CGFFCTEE255 pKa = 4.02NKK257 pKa = 10.3SAIQPDD263 pKa = 4.73LIEE266 pKa = 3.96VCARR270 pKa = 11.84VVFQMRR276 pKa = 11.84GRR278 pKa = 11.84KK279 pKa = 8.84NLGTEE284 pKa = 4.23TDD286 pKa = 3.09SSGAFDD292 pKa = 3.34AAEE295 pKa = 3.92VVEE298 pKa = 5.4FSMDD302 pKa = 3.37QRR304 pKa = 11.84GTAPPKK310 pKa = 10.32LRR312 pKa = 11.84YY313 pKa = 9.62GPLKK317 pKa = 10.46ALPDD321 pKa = 3.74CSSFKK326 pKa = 10.46TSNPTDD332 pKa = 3.38LHH334 pKa = 5.86VQQITCSS341 pKa = 3.4
Molecular weight: 39.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608 |
267 |
341 |
304.0 |
35.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 1.321 | 2.138 ± 0.3 |
3.783 ± 0.256 | 4.934 ± 1.121 |
4.605 ± 0.163 | 5.428 ± 1.279 |
1.645 ± 0.606 | 4.441 ± 0.498 |
5.428 ± 1.047 | 5.921 ± 0.277 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.151 ± 0.017 | 3.289 ± 0.05 |
5.757 ± 0.783 | 5.921 ± 0.741 |
9.046 ± 2.125 | 7.895 ± 1.179 |
6.579 ± 0.131 | 7.895 ± 1.412 |
3.783 ± 0.906 | 4.112 ± 0.237 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
