
Torque teno midi virus 11
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.28
Get precalculated fractions of proteins
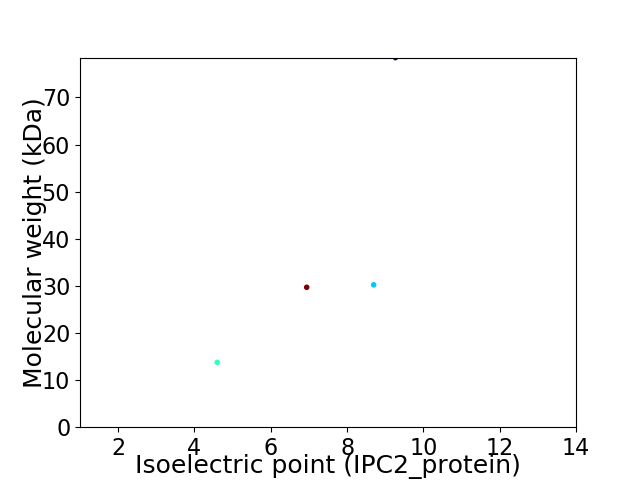
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7VLZ2|A7VLZ2_9VIRU Capsid protein OS=Torque teno midi virus 11 OX=2065052 PE=3 SV=1
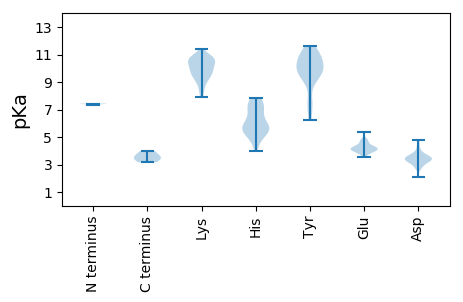
MM1 pKa = 7.43SNYY4 pKa = 9.79SANHH8 pKa = 5.58FFKK11 pKa = 9.81PTPYY15 pKa = 11.19NNDD18 pKa = 3.14TKK20 pKa = 10.99NQMWMSMISDD30 pKa = 3.76GHH32 pKa = 7.87DD33 pKa = 3.38CMCNCPSPFGHH44 pKa = 7.32LLASIFPPGHH54 pKa = 5.66QDD56 pKa = 2.75RR57 pKa = 11.84FLTVEE62 pKa = 4.18EE63 pKa = 4.78IINRR67 pKa = 11.84DD68 pKa = 2.84IKK70 pKa = 10.74EE71 pKa = 4.23CLFGGPDD78 pKa = 3.32ARR80 pKa = 11.84DD81 pKa = 3.54GGEE84 pKa = 3.83AAAGPSTGVDD94 pKa = 3.27TTIKK98 pKa = 10.65RR99 pKa = 11.84EE100 pKa = 4.03DD101 pKa = 3.66PEE103 pKa = 4.24EE104 pKa = 4.81DD105 pKa = 3.17FTKK108 pKa = 10.91EE109 pKa = 3.95NIEE112 pKa = 4.02EE113 pKa = 4.6LIAAAEE119 pKa = 4.24KK120 pKa = 10.61ASEE123 pKa = 4.08RR124 pKa = 3.99
MM1 pKa = 7.43SNYY4 pKa = 9.79SANHH8 pKa = 5.58FFKK11 pKa = 9.81PTPYY15 pKa = 11.19NNDD18 pKa = 3.14TKK20 pKa = 10.99NQMWMSMISDD30 pKa = 3.76GHH32 pKa = 7.87DD33 pKa = 3.38CMCNCPSPFGHH44 pKa = 7.32LLASIFPPGHH54 pKa = 5.66QDD56 pKa = 2.75RR57 pKa = 11.84FLTVEE62 pKa = 4.18EE63 pKa = 4.78IINRR67 pKa = 11.84DD68 pKa = 2.84IKK70 pKa = 10.74EE71 pKa = 4.23CLFGGPDD78 pKa = 3.32ARR80 pKa = 11.84DD81 pKa = 3.54GGEE84 pKa = 3.83AAAGPSTGVDD94 pKa = 3.27TTIKK98 pKa = 10.65RR99 pKa = 11.84EE100 pKa = 4.03DD101 pKa = 3.66PEE103 pKa = 4.24EE104 pKa = 4.81DD105 pKa = 3.17FTKK108 pKa = 10.91EE109 pKa = 3.95NIEE112 pKa = 4.02EE113 pKa = 4.6LIAAAEE119 pKa = 4.24KK120 pKa = 10.61ASEE123 pKa = 4.08RR124 pKa = 3.99
Molecular weight: 13.77 kDa
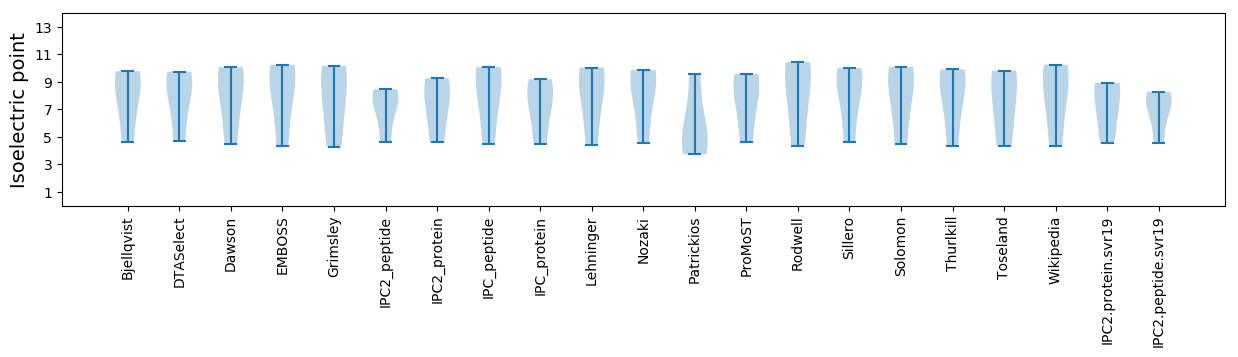
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLZ2|A7VLZ2_9VIRU Capsid protein OS=Torque teno midi virus 11 OX=2065052 PE=3 SV=1
MM1 pKa = 7.31PFWWARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.46RR10 pKa = 11.84WWRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PFYY20 pKa = 10.23RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.77HH25 pKa = 5.21YY26 pKa = 10.11KK27 pKa = 9.06KK28 pKa = 9.73RR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84LYY36 pKa = 10.04KK37 pKa = 9.27RR38 pKa = 11.84KK39 pKa = 9.35HH40 pKa = 5.07RR41 pKa = 11.84RR42 pKa = 11.84TYY44 pKa = 9.27RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.42SFRR52 pKa = 11.84KK53 pKa = 8.97VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 10.05RR58 pKa = 11.84RR59 pKa = 11.84TLPLKK64 pKa = 9.27QWQPEE69 pKa = 4.35TIRR72 pKa = 11.84KK73 pKa = 9.43CKK75 pKa = 10.14VKK77 pKa = 10.79GIGIHH82 pKa = 4.78TLGVEE87 pKa = 4.66GTQYY91 pKa = 11.03RR92 pKa = 11.84CYY94 pKa = 9.66TANKK98 pKa = 9.43YY99 pKa = 10.36EE100 pKa = 4.0YY101 pKa = 9.48TIAKK105 pKa = 8.75QPSGGGFGVEE115 pKa = 4.2RR116 pKa = 11.84YY117 pKa = 7.44TLEE120 pKa = 3.97YY121 pKa = 10.32LYY123 pKa = 10.79KK124 pKa = 10.04EE125 pKa = 4.3HH126 pKa = 7.66KK127 pKa = 10.28AGNNVWTTSNVNLDD141 pKa = 3.41LVRR144 pKa = 11.84YY145 pKa = 6.23TGCRR149 pKa = 11.84FIFWRR154 pKa = 11.84HH155 pKa = 3.98PHH157 pKa = 6.03IDD159 pKa = 3.41FVGVYY164 pKa = 9.66SRR166 pKa = 11.84NYY168 pKa = 9.26PMQLQKK174 pKa = 10.04YY175 pKa = 6.44TYY177 pKa = 9.49PDD179 pKa = 3.22THH181 pKa = 6.77PQQIMLAKK189 pKa = 9.53HH190 pKa = 5.82KK191 pKa = 10.46FFIPSLLTKK200 pKa = 9.7PHH202 pKa = 5.8GKK204 pKa = 9.77RR205 pKa = 11.84YY206 pKa = 10.29VKK208 pKa = 10.36IKK210 pKa = 10.27IKK212 pKa = 10.6PPTQMSNKK220 pKa = 8.79WFFQEE225 pKa = 4.48GFSDD229 pKa = 3.44TGLVQIHH236 pKa = 5.95TAACDD241 pKa = 3.23LRR243 pKa = 11.84YY244 pKa = 10.19AHH246 pKa = 7.32LGCCNTNQLASFLTLNTDD264 pKa = 4.78FYY266 pKa = 11.62QYY268 pKa = 11.27AAWGNPKK275 pKa = 10.07NPYY278 pKa = 9.92IPTEE282 pKa = 3.55QDD284 pKa = 2.06KK285 pKa = 10.0WYY287 pKa = 9.4RR288 pKa = 11.84PYY290 pKa = 10.81RR291 pKa = 11.84IVEE294 pKa = 4.09PVTEE298 pKa = 4.16VEE300 pKa = 4.31DD301 pKa = 3.55VKK303 pKa = 11.4GNKK306 pKa = 8.17KK307 pKa = 9.15TITTVFTPYY316 pKa = 10.51TEE318 pKa = 4.46TITYY322 pKa = 9.28NKK324 pKa = 10.24GWFQPALLQAKK335 pKa = 9.64QITKK339 pKa = 9.82PSQANPPLKK348 pKa = 9.76GARR351 pKa = 11.84YY352 pKa = 9.94NPTLDD357 pKa = 3.27TGEE360 pKa = 4.2NTAVWLTNVVNQSYY374 pKa = 9.61YY375 pKa = 10.04PPKK378 pKa = 9.68TDD380 pKa = 4.76LDD382 pKa = 4.28LFVQGLPLWQLLLGFPDD399 pKa = 3.51WVRR402 pKa = 11.84QKK404 pKa = 11.16KK405 pKa = 9.75KK406 pKa = 10.87DD407 pKa = 3.46PTFLQSYY414 pKa = 6.91YY415 pKa = 11.29LCFHH419 pKa = 7.43CDD421 pKa = 3.49AIEE424 pKa = 4.28PKK426 pKa = 10.19PGLHH430 pKa = 5.58QVFIPVDD437 pKa = 3.4YY438 pKa = 11.31SFVQGKK444 pKa = 9.68GPYY447 pKa = 9.92DD448 pKa = 3.56SLLSKK453 pKa = 9.67WDD455 pKa = 3.43NDD457 pKa = 2.88HH458 pKa = 7.25WYY460 pKa = 7.43PTLKK464 pKa = 10.55HH465 pKa = 5.72QLQTINSVVEE475 pKa = 3.95TGPFVPKK482 pKa = 10.55LDD484 pKa = 3.73TLKK487 pKa = 10.99NSTWEE492 pKa = 4.17LYY494 pKa = 8.96STYY497 pKa = 10.52TFYY500 pKa = 11.19FKK502 pKa = 10.85FGGASLPEE510 pKa = 5.08AEE512 pKa = 4.86TKK514 pKa = 10.87DD515 pKa = 3.49PATQGTHH522 pKa = 5.54EE523 pKa = 4.55VPDD526 pKa = 4.0KK527 pKa = 10.38QSKK530 pKa = 10.09AIQIINPKK538 pKa = 10.05KK539 pKa = 8.47NTAAAALHH547 pKa = 6.22CWDD550 pKa = 4.23FRR552 pKa = 11.84RR553 pKa = 11.84GHH555 pKa = 4.94ITSRR559 pKa = 11.84AFKK562 pKa = 10.37RR563 pKa = 11.84MCEE566 pKa = 3.88NAEE569 pKa = 3.98TDD571 pKa = 3.16SDD573 pKa = 3.98FQADD577 pKa = 3.72VSPKK581 pKa = 8.75KK582 pKa = 9.3TKK584 pKa = 10.01TSLQEE589 pKa = 3.68NTLPCISKK597 pKa = 10.05EE598 pKa = 3.97AEE600 pKa = 3.96EE601 pKa = 4.39VQDD604 pKa = 4.47CLQSLYY610 pKa = 11.03EE611 pKa = 4.15EE612 pKa = 4.37NTFQEE617 pKa = 5.35IPDD620 pKa = 3.66QQNIQQLLHH629 pKa = 5.65QQQQQQQHH637 pKa = 5.24TKK639 pKa = 10.42LQLLQLINNLKK650 pKa = 10.22KK651 pKa = 10.18KK652 pKa = 10.37QKK654 pKa = 10.39LIQLQTGILDD664 pKa = 3.51
MM1 pKa = 7.31PFWWARR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.46RR10 pKa = 11.84WWRR13 pKa = 11.84GRR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PFYY20 pKa = 10.23RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84YY24 pKa = 7.77HH25 pKa = 5.21YY26 pKa = 10.11KK27 pKa = 9.06KK28 pKa = 9.73RR29 pKa = 11.84RR30 pKa = 11.84PRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84LYY36 pKa = 10.04KK37 pKa = 9.27RR38 pKa = 11.84KK39 pKa = 9.35HH40 pKa = 5.07RR41 pKa = 11.84RR42 pKa = 11.84TYY44 pKa = 9.27RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 9.42SFRR52 pKa = 11.84KK53 pKa = 8.97VRR55 pKa = 11.84RR56 pKa = 11.84KK57 pKa = 10.05RR58 pKa = 11.84RR59 pKa = 11.84TLPLKK64 pKa = 9.27QWQPEE69 pKa = 4.35TIRR72 pKa = 11.84KK73 pKa = 9.43CKK75 pKa = 10.14VKK77 pKa = 10.79GIGIHH82 pKa = 4.78TLGVEE87 pKa = 4.66GTQYY91 pKa = 11.03RR92 pKa = 11.84CYY94 pKa = 9.66TANKK98 pKa = 9.43YY99 pKa = 10.36EE100 pKa = 4.0YY101 pKa = 9.48TIAKK105 pKa = 8.75QPSGGGFGVEE115 pKa = 4.2RR116 pKa = 11.84YY117 pKa = 7.44TLEE120 pKa = 3.97YY121 pKa = 10.32LYY123 pKa = 10.79KK124 pKa = 10.04EE125 pKa = 4.3HH126 pKa = 7.66KK127 pKa = 10.28AGNNVWTTSNVNLDD141 pKa = 3.41LVRR144 pKa = 11.84YY145 pKa = 6.23TGCRR149 pKa = 11.84FIFWRR154 pKa = 11.84HH155 pKa = 3.98PHH157 pKa = 6.03IDD159 pKa = 3.41FVGVYY164 pKa = 9.66SRR166 pKa = 11.84NYY168 pKa = 9.26PMQLQKK174 pKa = 10.04YY175 pKa = 6.44TYY177 pKa = 9.49PDD179 pKa = 3.22THH181 pKa = 6.77PQQIMLAKK189 pKa = 9.53HH190 pKa = 5.82KK191 pKa = 10.46FFIPSLLTKK200 pKa = 9.7PHH202 pKa = 5.8GKK204 pKa = 9.77RR205 pKa = 11.84YY206 pKa = 10.29VKK208 pKa = 10.36IKK210 pKa = 10.27IKK212 pKa = 10.6PPTQMSNKK220 pKa = 8.79WFFQEE225 pKa = 4.48GFSDD229 pKa = 3.44TGLVQIHH236 pKa = 5.95TAACDD241 pKa = 3.23LRR243 pKa = 11.84YY244 pKa = 10.19AHH246 pKa = 7.32LGCCNTNQLASFLTLNTDD264 pKa = 4.78FYY266 pKa = 11.62QYY268 pKa = 11.27AAWGNPKK275 pKa = 10.07NPYY278 pKa = 9.92IPTEE282 pKa = 3.55QDD284 pKa = 2.06KK285 pKa = 10.0WYY287 pKa = 9.4RR288 pKa = 11.84PYY290 pKa = 10.81RR291 pKa = 11.84IVEE294 pKa = 4.09PVTEE298 pKa = 4.16VEE300 pKa = 4.31DD301 pKa = 3.55VKK303 pKa = 11.4GNKK306 pKa = 8.17KK307 pKa = 9.15TITTVFTPYY316 pKa = 10.51TEE318 pKa = 4.46TITYY322 pKa = 9.28NKK324 pKa = 10.24GWFQPALLQAKK335 pKa = 9.64QITKK339 pKa = 9.82PSQANPPLKK348 pKa = 9.76GARR351 pKa = 11.84YY352 pKa = 9.94NPTLDD357 pKa = 3.27TGEE360 pKa = 4.2NTAVWLTNVVNQSYY374 pKa = 9.61YY375 pKa = 10.04PPKK378 pKa = 9.68TDD380 pKa = 4.76LDD382 pKa = 4.28LFVQGLPLWQLLLGFPDD399 pKa = 3.51WVRR402 pKa = 11.84QKK404 pKa = 11.16KK405 pKa = 9.75KK406 pKa = 10.87DD407 pKa = 3.46PTFLQSYY414 pKa = 6.91YY415 pKa = 11.29LCFHH419 pKa = 7.43CDD421 pKa = 3.49AIEE424 pKa = 4.28PKK426 pKa = 10.19PGLHH430 pKa = 5.58QVFIPVDD437 pKa = 3.4YY438 pKa = 11.31SFVQGKK444 pKa = 9.68GPYY447 pKa = 9.92DD448 pKa = 3.56SLLSKK453 pKa = 9.67WDD455 pKa = 3.43NDD457 pKa = 2.88HH458 pKa = 7.25WYY460 pKa = 7.43PTLKK464 pKa = 10.55HH465 pKa = 5.72QLQTINSVVEE475 pKa = 3.95TGPFVPKK482 pKa = 10.55LDD484 pKa = 3.73TLKK487 pKa = 10.99NSTWEE492 pKa = 4.17LYY494 pKa = 8.96STYY497 pKa = 10.52TFYY500 pKa = 11.19FKK502 pKa = 10.85FGGASLPEE510 pKa = 5.08AEE512 pKa = 4.86TKK514 pKa = 10.87DD515 pKa = 3.49PATQGTHH522 pKa = 5.54EE523 pKa = 4.55VPDD526 pKa = 4.0KK527 pKa = 10.38QSKK530 pKa = 10.09AIQIINPKK538 pKa = 10.05KK539 pKa = 8.47NTAAAALHH547 pKa = 6.22CWDD550 pKa = 4.23FRR552 pKa = 11.84RR553 pKa = 11.84GHH555 pKa = 4.94ITSRR559 pKa = 11.84AFKK562 pKa = 10.37RR563 pKa = 11.84MCEE566 pKa = 3.88NAEE569 pKa = 3.98TDD571 pKa = 3.16SDD573 pKa = 3.98FQADD577 pKa = 3.72VSPKK581 pKa = 8.75KK582 pKa = 9.3TKK584 pKa = 10.01TSLQEE589 pKa = 3.68NTLPCISKK597 pKa = 10.05EE598 pKa = 3.97AEE600 pKa = 3.96EE601 pKa = 4.39VQDD604 pKa = 4.47CLQSLYY610 pKa = 11.03EE611 pKa = 4.15EE612 pKa = 4.37NTFQEE617 pKa = 5.35IPDD620 pKa = 3.66QQNIQQLLHH629 pKa = 5.65QQQQQQQHH637 pKa = 5.24TKK639 pKa = 10.42LQLLQLINNLKK650 pKa = 10.22KK651 pKa = 10.18KK652 pKa = 10.37QKK654 pKa = 10.39LIQLQTGILDD664 pKa = 3.51
Molecular weight: 78.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1321 |
124 |
664 |
330.3 |
38.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.98 ± 1.204 | 2.044 ± 0.214 |
4.466 ± 0.511 | 6.056 ± 1.244 |
4.391 ± 0.406 | 4.769 ± 0.368 |
3.482 ± 0.355 | 4.542 ± 0.276 |
8.554 ± 0.945 | 6.889 ± 1.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.741 ± 0.679 | 4.921 ± 0.443 |
7.419 ± 1.184 | 5.829 ± 1.283 |
5.829 ± 0.696 | 6.435 ± 2.421 |
7.873 ± 0.423 | 3.104 ± 0.928 |
1.665 ± 0.698 | 4.012 ± 1.137 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
