
Tobacco vein clearing virus (isolate Nicotiana edwardsonii/United States/Lockhart/2000) (TVCV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Solendovirus; Tobacco vein clearing virus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
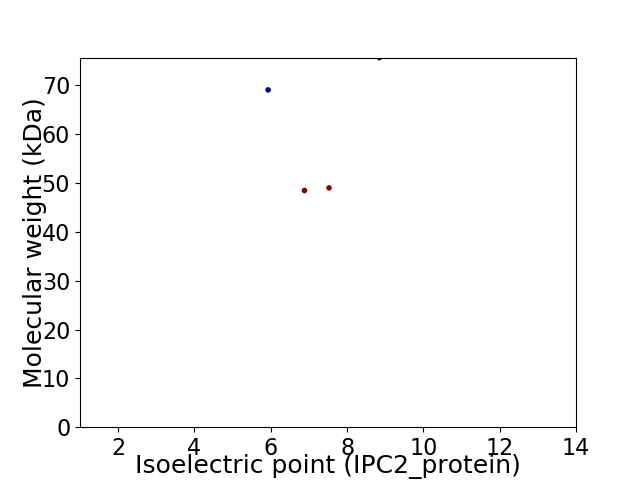
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9QD06|Q9QD06_TVCVL Coat protein OS=Tobacco vein clearing virus (isolate Nicotiana edwardsonii/United States/Lockhart/2000) OX=1289468 PE=3 SV=1
MM1 pKa = 7.29NKK3 pKa = 9.92EE4 pKa = 3.58EE5 pKa = 3.95FAIDD9 pKa = 3.23EE10 pKa = 4.39KK11 pKa = 10.6TYY13 pKa = 10.94EE14 pKa = 4.17NQDD17 pKa = 3.09GLKK20 pKa = 10.16IKK22 pKa = 10.43IIFSNLGRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 10.28KK33 pKa = 10.75KK34 pKa = 10.44IGDD37 pKa = 3.75QIYY40 pKa = 11.08LMIEE44 pKa = 3.89KK45 pKa = 8.47EE46 pKa = 4.23TARR49 pKa = 11.84LEE51 pKa = 4.41DD52 pKa = 3.99SLTAMTRR59 pKa = 11.84IIKK62 pKa = 10.09EE63 pKa = 3.74NEE65 pKa = 3.9EE66 pKa = 3.59IDD68 pKa = 3.41KK69 pKa = 10.88KK70 pKa = 11.12RR71 pKa = 11.84EE72 pKa = 3.47IEE74 pKa = 4.3IIRR77 pKa = 11.84NQANKK82 pKa = 10.06EE83 pKa = 3.92IQQLEE88 pKa = 4.2EE89 pKa = 4.08TKK91 pKa = 9.21NTKK94 pKa = 10.06IIALEE99 pKa = 4.22KK100 pKa = 9.27EE101 pKa = 4.24LNMLKK106 pKa = 10.56LLYY109 pKa = 10.1EE110 pKa = 4.21NKK112 pKa = 9.44QRR114 pKa = 11.84EE115 pKa = 4.03KK116 pKa = 11.04DD117 pKa = 3.63KK118 pKa = 11.07EE119 pKa = 3.95IEE121 pKa = 4.12LTDD124 pKa = 4.33EE125 pKa = 3.92INKK128 pKa = 9.34FKK130 pKa = 11.04QKK132 pKa = 10.41LQPEE136 pKa = 4.19EE137 pKa = 3.68NLRR140 pKa = 11.84IEE142 pKa = 5.14EE143 pKa = 4.08IDD145 pKa = 4.68NNIDD149 pKa = 3.58DD150 pKa = 4.38QKK152 pKa = 11.6SEE154 pKa = 3.98LSEE157 pKa = 3.73ISEE160 pKa = 4.7TYY162 pKa = 10.27TEE164 pKa = 3.8ILEE167 pKa = 4.7NIEE170 pKa = 3.84KK171 pKa = 9.9TKK173 pKa = 10.74NLEE176 pKa = 3.93INTGDD181 pKa = 3.23VNMDD185 pKa = 4.01EE186 pKa = 4.93KK187 pKa = 10.97PSTSGIKK194 pKa = 9.88NPKK197 pKa = 8.41EE198 pKa = 4.03LNPSYY203 pKa = 9.95YY204 pKa = 9.19TVSYY208 pKa = 9.33EE209 pKa = 4.24QSDD212 pKa = 3.94RR213 pKa = 11.84NNTLWNSRR221 pKa = 11.84LNKK224 pKa = 9.73KK225 pKa = 6.57WTPKK229 pKa = 9.99PIHH232 pKa = 5.59EE233 pKa = 4.55QYY235 pKa = 11.56NFLDD239 pKa = 4.21LDD241 pKa = 3.99CVEE244 pKa = 5.83DD245 pKa = 3.67INKK248 pKa = 8.83TIQLWIGYY256 pKa = 9.01ISKK259 pKa = 10.16QLIDD263 pKa = 3.65NKK265 pKa = 10.46IGITEE270 pKa = 4.03TPGYY274 pKa = 9.94IEE276 pKa = 4.09RR277 pKa = 11.84TLIGTVKK284 pKa = 10.38LWLHH288 pKa = 6.09NLTKK292 pKa = 10.71EE293 pKa = 4.47SIDD296 pKa = 3.44TLRR299 pKa = 11.84SNKK302 pKa = 9.39KK303 pKa = 10.28FNGDD307 pKa = 3.05MATTSMEE314 pKa = 3.66ILYY317 pKa = 9.84KK318 pKa = 10.41YY319 pKa = 9.98EE320 pKa = 3.44IAIRR324 pKa = 11.84NEE326 pKa = 3.89FSSMTTEE333 pKa = 3.81TEE335 pKa = 3.98EE336 pKa = 3.8QHH338 pKa = 7.01KK339 pKa = 10.21EE340 pKa = 3.64KK341 pKa = 10.73QINRR345 pKa = 11.84NLMTKK350 pKa = 10.48LAICNMCYY358 pKa = 9.36IDD360 pKa = 5.37EE361 pKa = 4.4YY362 pKa = 9.24TCAFRR367 pKa = 11.84EE368 pKa = 4.21YY369 pKa = 10.12YY370 pKa = 10.55YY371 pKa = 10.95KK372 pKa = 10.25GTYY375 pKa = 7.49NTEE378 pKa = 3.68EE379 pKa = 3.82GRR381 pKa = 11.84EE382 pKa = 3.86IRR384 pKa = 11.84KK385 pKa = 9.8LYY387 pKa = 8.11FTKK390 pKa = 10.57LPEE393 pKa = 4.44PFSSKK398 pKa = 10.1IIKK401 pKa = 9.51DD402 pKa = 3.25WNEE405 pKa = 3.56AGLTDD410 pKa = 3.92TLGARR415 pKa = 11.84IKK417 pKa = 10.46FLQQWFVQLCEE428 pKa = 4.25KK429 pKa = 10.53YY430 pKa = 9.85KK431 pKa = 11.07EE432 pKa = 4.1EE433 pKa = 4.01IKK435 pKa = 10.1MEE437 pKa = 4.81KK438 pKa = 9.95ILIKK442 pKa = 10.5NLKK445 pKa = 8.3CCKK448 pKa = 10.33NKK450 pKa = 8.96TAPQFGCTDD459 pKa = 3.66KK460 pKa = 11.55YY461 pKa = 9.57NKK463 pKa = 9.24KK464 pKa = 9.59WKK466 pKa = 7.12TKK468 pKa = 9.42KK469 pKa = 9.74YY470 pKa = 8.88KK471 pKa = 9.96KK472 pKa = 9.68HH473 pKa = 5.88RR474 pKa = 11.84SKK476 pKa = 11.29YY477 pKa = 9.46KK478 pKa = 8.9YY479 pKa = 8.76KK480 pKa = 10.52KK481 pKa = 8.41PRR483 pKa = 11.84KK484 pKa = 8.48RR485 pKa = 11.84YY486 pKa = 8.5YY487 pKa = 9.67VKK489 pKa = 10.52NYY491 pKa = 7.26QAKK494 pKa = 10.24KK495 pKa = 9.49PFRR498 pKa = 11.84PKK500 pKa = 10.55KK501 pKa = 10.26KK502 pKa = 9.27LTEE505 pKa = 4.2CTCYY509 pKa = 10.48NCGKK513 pKa = 9.63LGHH516 pKa = 6.75IAKK519 pKa = 9.97DD520 pKa = 3.64CKK522 pKa = 10.8APKK525 pKa = 9.88NPKK528 pKa = 9.38KK529 pKa = 10.6KK530 pKa = 10.33QITEE534 pKa = 4.1IIIDD538 pKa = 3.97DD539 pKa = 4.18EE540 pKa = 5.3EE541 pKa = 4.68YY542 pKa = 10.36MQMEE546 pKa = 4.74YY547 pKa = 9.93IDD549 pKa = 4.54YY550 pKa = 10.44EE551 pKa = 4.42LDD553 pKa = 3.76SEE555 pKa = 4.4DD556 pKa = 4.11SIYY559 pKa = 10.68EE560 pKa = 3.79ISDD563 pKa = 3.3SDD565 pKa = 4.76DD566 pKa = 4.7EE567 pKa = 5.01IDD569 pKa = 4.38SEE571 pKa = 4.22ITEE574 pKa = 4.4EE575 pKa = 3.96EE576 pKa = 4.37DD577 pKa = 4.17EE578 pKa = 4.27II579 pKa = 6.24
MM1 pKa = 7.29NKK3 pKa = 9.92EE4 pKa = 3.58EE5 pKa = 3.95FAIDD9 pKa = 3.23EE10 pKa = 4.39KK11 pKa = 10.6TYY13 pKa = 10.94EE14 pKa = 4.17NQDD17 pKa = 3.09GLKK20 pKa = 10.16IKK22 pKa = 10.43IIFSNLGRR30 pKa = 11.84RR31 pKa = 11.84YY32 pKa = 10.28KK33 pKa = 10.75KK34 pKa = 10.44IGDD37 pKa = 3.75QIYY40 pKa = 11.08LMIEE44 pKa = 3.89KK45 pKa = 8.47EE46 pKa = 4.23TARR49 pKa = 11.84LEE51 pKa = 4.41DD52 pKa = 3.99SLTAMTRR59 pKa = 11.84IIKK62 pKa = 10.09EE63 pKa = 3.74NEE65 pKa = 3.9EE66 pKa = 3.59IDD68 pKa = 3.41KK69 pKa = 10.88KK70 pKa = 11.12RR71 pKa = 11.84EE72 pKa = 3.47IEE74 pKa = 4.3IIRR77 pKa = 11.84NQANKK82 pKa = 10.06EE83 pKa = 3.92IQQLEE88 pKa = 4.2EE89 pKa = 4.08TKK91 pKa = 9.21NTKK94 pKa = 10.06IIALEE99 pKa = 4.22KK100 pKa = 9.27EE101 pKa = 4.24LNMLKK106 pKa = 10.56LLYY109 pKa = 10.1EE110 pKa = 4.21NKK112 pKa = 9.44QRR114 pKa = 11.84EE115 pKa = 4.03KK116 pKa = 11.04DD117 pKa = 3.63KK118 pKa = 11.07EE119 pKa = 3.95IEE121 pKa = 4.12LTDD124 pKa = 4.33EE125 pKa = 3.92INKK128 pKa = 9.34FKK130 pKa = 11.04QKK132 pKa = 10.41LQPEE136 pKa = 4.19EE137 pKa = 3.68NLRR140 pKa = 11.84IEE142 pKa = 5.14EE143 pKa = 4.08IDD145 pKa = 4.68NNIDD149 pKa = 3.58DD150 pKa = 4.38QKK152 pKa = 11.6SEE154 pKa = 3.98LSEE157 pKa = 3.73ISEE160 pKa = 4.7TYY162 pKa = 10.27TEE164 pKa = 3.8ILEE167 pKa = 4.7NIEE170 pKa = 3.84KK171 pKa = 9.9TKK173 pKa = 10.74NLEE176 pKa = 3.93INTGDD181 pKa = 3.23VNMDD185 pKa = 4.01EE186 pKa = 4.93KK187 pKa = 10.97PSTSGIKK194 pKa = 9.88NPKK197 pKa = 8.41EE198 pKa = 4.03LNPSYY203 pKa = 9.95YY204 pKa = 9.19TVSYY208 pKa = 9.33EE209 pKa = 4.24QSDD212 pKa = 3.94RR213 pKa = 11.84NNTLWNSRR221 pKa = 11.84LNKK224 pKa = 9.73KK225 pKa = 6.57WTPKK229 pKa = 9.99PIHH232 pKa = 5.59EE233 pKa = 4.55QYY235 pKa = 11.56NFLDD239 pKa = 4.21LDD241 pKa = 3.99CVEE244 pKa = 5.83DD245 pKa = 3.67INKK248 pKa = 8.83TIQLWIGYY256 pKa = 9.01ISKK259 pKa = 10.16QLIDD263 pKa = 3.65NKK265 pKa = 10.46IGITEE270 pKa = 4.03TPGYY274 pKa = 9.94IEE276 pKa = 4.09RR277 pKa = 11.84TLIGTVKK284 pKa = 10.38LWLHH288 pKa = 6.09NLTKK292 pKa = 10.71EE293 pKa = 4.47SIDD296 pKa = 3.44TLRR299 pKa = 11.84SNKK302 pKa = 9.39KK303 pKa = 10.28FNGDD307 pKa = 3.05MATTSMEE314 pKa = 3.66ILYY317 pKa = 9.84KK318 pKa = 10.41YY319 pKa = 9.98EE320 pKa = 3.44IAIRR324 pKa = 11.84NEE326 pKa = 3.89FSSMTTEE333 pKa = 3.81TEE335 pKa = 3.98EE336 pKa = 3.8QHH338 pKa = 7.01KK339 pKa = 10.21EE340 pKa = 3.64KK341 pKa = 10.73QINRR345 pKa = 11.84NLMTKK350 pKa = 10.48LAICNMCYY358 pKa = 9.36IDD360 pKa = 5.37EE361 pKa = 4.4YY362 pKa = 9.24TCAFRR367 pKa = 11.84EE368 pKa = 4.21YY369 pKa = 10.12YY370 pKa = 10.55YY371 pKa = 10.95KK372 pKa = 10.25GTYY375 pKa = 7.49NTEE378 pKa = 3.68EE379 pKa = 3.82GRR381 pKa = 11.84EE382 pKa = 3.86IRR384 pKa = 11.84KK385 pKa = 9.8LYY387 pKa = 8.11FTKK390 pKa = 10.57LPEE393 pKa = 4.44PFSSKK398 pKa = 10.1IIKK401 pKa = 9.51DD402 pKa = 3.25WNEE405 pKa = 3.56AGLTDD410 pKa = 3.92TLGARR415 pKa = 11.84IKK417 pKa = 10.46FLQQWFVQLCEE428 pKa = 4.25KK429 pKa = 10.53YY430 pKa = 9.85KK431 pKa = 11.07EE432 pKa = 4.1EE433 pKa = 4.01IKK435 pKa = 10.1MEE437 pKa = 4.81KK438 pKa = 9.95ILIKK442 pKa = 10.5NLKK445 pKa = 8.3CCKK448 pKa = 10.33NKK450 pKa = 8.96TAPQFGCTDD459 pKa = 3.66KK460 pKa = 11.55YY461 pKa = 9.57NKK463 pKa = 9.24KK464 pKa = 9.59WKK466 pKa = 7.12TKK468 pKa = 9.42KK469 pKa = 9.74YY470 pKa = 8.88KK471 pKa = 9.96KK472 pKa = 9.68HH473 pKa = 5.88RR474 pKa = 11.84SKK476 pKa = 11.29YY477 pKa = 9.46KK478 pKa = 8.9YY479 pKa = 8.76KK480 pKa = 10.52KK481 pKa = 8.41PRR483 pKa = 11.84KK484 pKa = 8.48RR485 pKa = 11.84YY486 pKa = 8.5YY487 pKa = 9.67VKK489 pKa = 10.52NYY491 pKa = 7.26QAKK494 pKa = 10.24KK495 pKa = 9.49PFRR498 pKa = 11.84PKK500 pKa = 10.55KK501 pKa = 10.26KK502 pKa = 9.27LTEE505 pKa = 4.2CTCYY509 pKa = 10.48NCGKK513 pKa = 9.63LGHH516 pKa = 6.75IAKK519 pKa = 9.97DD520 pKa = 3.64CKK522 pKa = 10.8APKK525 pKa = 9.88NPKK528 pKa = 9.38KK529 pKa = 10.6KK530 pKa = 10.33QITEE534 pKa = 4.1IIIDD538 pKa = 3.97DD539 pKa = 4.18EE540 pKa = 5.3EE541 pKa = 4.68YY542 pKa = 10.36MQMEE546 pKa = 4.74YY547 pKa = 9.93IDD549 pKa = 4.54YY550 pKa = 10.44EE551 pKa = 4.42LDD553 pKa = 3.76SEE555 pKa = 4.4DD556 pKa = 4.11SIYY559 pKa = 10.68EE560 pKa = 3.79ISDD563 pKa = 3.3SDD565 pKa = 4.76DD566 pKa = 4.7EE567 pKa = 5.01IDD569 pKa = 4.38SEE571 pKa = 4.22ITEE574 pKa = 4.4EE575 pKa = 3.96EE576 pKa = 4.37DD577 pKa = 4.17EE578 pKa = 4.27II579 pKa = 6.24
Molecular weight: 69.08 kDa
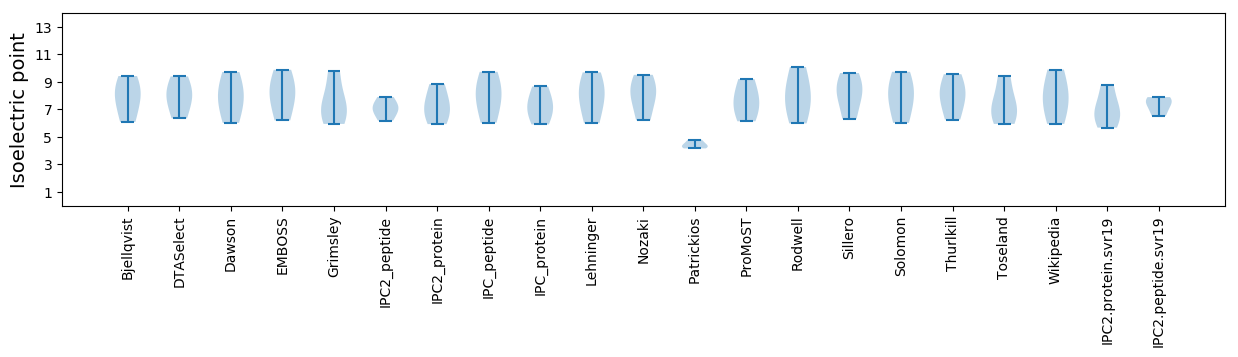
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9QD05|Q9QD05_TVCVL Putative cell-to-cell movement protein OS=Tobacco vein clearing virus (isolate Nicotiana edwardsonii/United States/Lockhart/2000) OX=1289468 PE=4 SV=1
MM1 pKa = 7.76KK2 pKa = 9.95IYY4 pKa = 10.16ILAKK8 pKa = 9.84IIVEE12 pKa = 4.68GYY14 pKa = 7.64YY15 pKa = 10.52NRR17 pKa = 11.84YY18 pKa = 5.62YY19 pKa = 10.91TPMIDD24 pKa = 3.21TGAEE28 pKa = 3.94ANICKK33 pKa = 10.01YY34 pKa = 10.96NCLPTDD40 pKa = 2.7KK41 pKa = 10.12WEE43 pKa = 4.11KK44 pKa = 10.57LKK46 pKa = 10.39TPMVVTGFNNEE57 pKa = 3.55GSMINYY63 pKa = 8.05KK64 pKa = 10.3ARR66 pKa = 11.84NVKK69 pKa = 9.16IQIWDD74 pKa = 3.82KK75 pKa = 10.56ILTIEE80 pKa = 4.47EE81 pKa = 4.27IYY83 pKa = 11.11NFEE86 pKa = 4.19FTTKK90 pKa = 10.94DD91 pKa = 3.28MLLGMPFLEE100 pKa = 4.44KK101 pKa = 10.31LYY103 pKa = 10.18PHH105 pKa = 7.77IITKK109 pKa = 5.99THH111 pKa = 4.35WWFTTPCKK119 pKa = 10.68NKK121 pKa = 10.06VGAKK125 pKa = 9.31RR126 pKa = 11.84VNNKK130 pKa = 7.37QRR132 pKa = 11.84KK133 pKa = 4.02TTEE136 pKa = 3.99WIRR139 pKa = 11.84GSEE142 pKa = 4.29KK143 pKa = 9.4ITQKK147 pKa = 11.04LEE149 pKa = 4.36NINKK153 pKa = 7.77NTTTQLEE160 pKa = 4.48IIIFTIDD167 pKa = 3.05KK168 pKa = 10.8VKK170 pKa = 10.49IIQKK174 pKa = 10.36KK175 pKa = 9.55LEE177 pKa = 3.98KK178 pKa = 10.37LYY180 pKa = 11.06NDD182 pKa = 4.09NPLQGWEE189 pKa = 3.85KK190 pKa = 10.67HH191 pKa = 4.0KK192 pKa = 9.91TKK194 pKa = 10.89VKK196 pKa = 10.4IEE198 pKa = 4.45LIEE201 pKa = 4.25EE202 pKa = 3.95NSIITQKK209 pKa = 9.58PLKK212 pKa = 10.73YY213 pKa = 10.34NFNDD217 pKa = 3.07LTEE220 pKa = 4.39FKK222 pKa = 10.57IHH224 pKa = 7.21IKK226 pKa = 10.67DD227 pKa = 3.75LLDD230 pKa = 3.42NKK232 pKa = 10.22YY233 pKa = 10.03IQEE236 pKa = 4.38SNSKK240 pKa = 8.07HH241 pKa = 5.22TSPAFIVNKK250 pKa = 10.11HH251 pKa = 5.18SEE253 pKa = 4.14QKK255 pKa = 9.79RR256 pKa = 11.84GKK258 pKa = 9.08SRR260 pKa = 11.84MVIDD264 pKa = 4.04YY265 pKa = 10.56RR266 pKa = 11.84NLNAKK271 pKa = 7.98TKK273 pKa = 8.87TYY275 pKa = 10.75NYY277 pKa = 7.77PIPNKK282 pKa = 9.55ILKK285 pKa = 9.19IRR287 pKa = 11.84QIQGYY292 pKa = 9.56NYY294 pKa = 9.96FSKK297 pKa = 10.61FDD299 pKa = 4.02CKK301 pKa = 10.9SGFYY305 pKa = 10.02HH306 pKa = 7.35LKK308 pKa = 10.93LEE310 pKa = 4.62DD311 pKa = 3.65EE312 pKa = 4.66SKK314 pKa = 11.06KK315 pKa = 9.77LTAFTVPQGFYY326 pKa = 9.41EE327 pKa = 4.28WNVLPFGYY335 pKa = 10.23KK336 pKa = 9.53NAPGRR341 pKa = 11.84YY342 pKa = 6.04QHH344 pKa = 6.91FMDD347 pKa = 4.7NYY349 pKa = 9.34FNQLEE354 pKa = 4.15NCIVYY359 pKa = 10.02IDD361 pKa = 5.64DD362 pKa = 3.36ILLYY366 pKa = 10.93SRR368 pKa = 11.84TQDD371 pKa = 3.14EE372 pKa = 4.81HH373 pKa = 7.93IKK375 pKa = 10.58LLEE378 pKa = 4.27KK379 pKa = 10.0FAHH382 pKa = 6.54IIEE385 pKa = 4.4NSGISLSKK393 pKa = 9.83TKK395 pKa = 11.0AEE397 pKa = 3.73IMKK400 pKa = 10.06NQIEE404 pKa = 4.17FLGIQIDD411 pKa = 3.82KK412 pKa = 11.11NGIKK416 pKa = 8.84MQTHH420 pKa = 6.44IVQKK424 pKa = 10.46IINLDD429 pKa = 3.6EE430 pKa = 5.32NIDD433 pKa = 3.95TKK435 pKa = 11.26KK436 pKa = 10.64KK437 pKa = 9.4LQSFLGIVNQVRR449 pKa = 11.84EE450 pKa = 4.78YY451 pKa = 10.52IPKK454 pKa = 9.98LAEE457 pKa = 3.88NLKK460 pKa = 10.03PLQKK464 pKa = 10.38KK465 pKa = 9.34LKK467 pKa = 10.02KK468 pKa = 10.1DD469 pKa = 3.28VEE471 pKa = 4.3YY472 pKa = 11.37SFDD475 pKa = 3.92EE476 pKa = 4.36KK477 pKa = 11.22DD478 pKa = 3.05KK479 pKa = 11.27EE480 pKa = 4.13QIKK483 pKa = 10.16KK484 pKa = 10.37IKK486 pKa = 9.72ILCKK490 pKa = 10.23KK491 pKa = 10.06LPKK494 pKa = 10.31LYY496 pKa = 10.72FPDD499 pKa = 3.63EE500 pKa = 4.39NKK502 pKa = 10.17KK503 pKa = 8.68FTYY506 pKa = 9.92IVEE509 pKa = 4.44TDD511 pKa = 3.28SSNYY515 pKa = 10.37SYY517 pKa = 11.63GGVLKK522 pKa = 10.7YY523 pKa = 10.35RR524 pKa = 11.84YY525 pKa = 8.69NKK527 pKa = 9.98EE528 pKa = 4.13KK529 pKa = 10.28IEE531 pKa = 3.99HH532 pKa = 5.78HH533 pKa = 6.1CRR535 pKa = 11.84YY536 pKa = 10.05YY537 pKa = 10.68SGSYY541 pKa = 9.4TEE543 pKa = 4.76PQEE546 pKa = 3.65KK547 pKa = 9.45WEE549 pKa = 4.18INRR552 pKa = 11.84KK553 pKa = 8.0EE554 pKa = 4.06LFALYY559 pKa = 10.07KK560 pKa = 10.4CLLAFEE566 pKa = 4.93PYY568 pKa = 9.5IVYY571 pKa = 9.05TRR573 pKa = 11.84FIVRR577 pKa = 11.84TDD579 pKa = 3.16NTQVKK584 pKa = 7.71WWITRR589 pKa = 11.84KK590 pKa = 9.67VQDD593 pKa = 3.37SVTTKK598 pKa = 10.13EE599 pKa = 3.62IRR601 pKa = 11.84RR602 pKa = 11.84LVLNILNFTFTIEE615 pKa = 4.96IINTNKK621 pKa = 10.13NVVADD626 pKa = 3.77YY627 pKa = 10.86LSRR630 pKa = 11.84QSYY633 pKa = 9.84PNN635 pKa = 3.3
MM1 pKa = 7.76KK2 pKa = 9.95IYY4 pKa = 10.16ILAKK8 pKa = 9.84IIVEE12 pKa = 4.68GYY14 pKa = 7.64YY15 pKa = 10.52NRR17 pKa = 11.84YY18 pKa = 5.62YY19 pKa = 10.91TPMIDD24 pKa = 3.21TGAEE28 pKa = 3.94ANICKK33 pKa = 10.01YY34 pKa = 10.96NCLPTDD40 pKa = 2.7KK41 pKa = 10.12WEE43 pKa = 4.11KK44 pKa = 10.57LKK46 pKa = 10.39TPMVVTGFNNEE57 pKa = 3.55GSMINYY63 pKa = 8.05KK64 pKa = 10.3ARR66 pKa = 11.84NVKK69 pKa = 9.16IQIWDD74 pKa = 3.82KK75 pKa = 10.56ILTIEE80 pKa = 4.47EE81 pKa = 4.27IYY83 pKa = 11.11NFEE86 pKa = 4.19FTTKK90 pKa = 10.94DD91 pKa = 3.28MLLGMPFLEE100 pKa = 4.44KK101 pKa = 10.31LYY103 pKa = 10.18PHH105 pKa = 7.77IITKK109 pKa = 5.99THH111 pKa = 4.35WWFTTPCKK119 pKa = 10.68NKK121 pKa = 10.06VGAKK125 pKa = 9.31RR126 pKa = 11.84VNNKK130 pKa = 7.37QRR132 pKa = 11.84KK133 pKa = 4.02TTEE136 pKa = 3.99WIRR139 pKa = 11.84GSEE142 pKa = 4.29KK143 pKa = 9.4ITQKK147 pKa = 11.04LEE149 pKa = 4.36NINKK153 pKa = 7.77NTTTQLEE160 pKa = 4.48IIIFTIDD167 pKa = 3.05KK168 pKa = 10.8VKK170 pKa = 10.49IIQKK174 pKa = 10.36KK175 pKa = 9.55LEE177 pKa = 3.98KK178 pKa = 10.37LYY180 pKa = 11.06NDD182 pKa = 4.09NPLQGWEE189 pKa = 3.85KK190 pKa = 10.67HH191 pKa = 4.0KK192 pKa = 9.91TKK194 pKa = 10.89VKK196 pKa = 10.4IEE198 pKa = 4.45LIEE201 pKa = 4.25EE202 pKa = 3.95NSIITQKK209 pKa = 9.58PLKK212 pKa = 10.73YY213 pKa = 10.34NFNDD217 pKa = 3.07LTEE220 pKa = 4.39FKK222 pKa = 10.57IHH224 pKa = 7.21IKK226 pKa = 10.67DD227 pKa = 3.75LLDD230 pKa = 3.42NKK232 pKa = 10.22YY233 pKa = 10.03IQEE236 pKa = 4.38SNSKK240 pKa = 8.07HH241 pKa = 5.22TSPAFIVNKK250 pKa = 10.11HH251 pKa = 5.18SEE253 pKa = 4.14QKK255 pKa = 9.79RR256 pKa = 11.84GKK258 pKa = 9.08SRR260 pKa = 11.84MVIDD264 pKa = 4.04YY265 pKa = 10.56RR266 pKa = 11.84NLNAKK271 pKa = 7.98TKK273 pKa = 8.87TYY275 pKa = 10.75NYY277 pKa = 7.77PIPNKK282 pKa = 9.55ILKK285 pKa = 9.19IRR287 pKa = 11.84QIQGYY292 pKa = 9.56NYY294 pKa = 9.96FSKK297 pKa = 10.61FDD299 pKa = 4.02CKK301 pKa = 10.9SGFYY305 pKa = 10.02HH306 pKa = 7.35LKK308 pKa = 10.93LEE310 pKa = 4.62DD311 pKa = 3.65EE312 pKa = 4.66SKK314 pKa = 11.06KK315 pKa = 9.77LTAFTVPQGFYY326 pKa = 9.41EE327 pKa = 4.28WNVLPFGYY335 pKa = 10.23KK336 pKa = 9.53NAPGRR341 pKa = 11.84YY342 pKa = 6.04QHH344 pKa = 6.91FMDD347 pKa = 4.7NYY349 pKa = 9.34FNQLEE354 pKa = 4.15NCIVYY359 pKa = 10.02IDD361 pKa = 5.64DD362 pKa = 3.36ILLYY366 pKa = 10.93SRR368 pKa = 11.84TQDD371 pKa = 3.14EE372 pKa = 4.81HH373 pKa = 7.93IKK375 pKa = 10.58LLEE378 pKa = 4.27KK379 pKa = 10.0FAHH382 pKa = 6.54IIEE385 pKa = 4.4NSGISLSKK393 pKa = 9.83TKK395 pKa = 11.0AEE397 pKa = 3.73IMKK400 pKa = 10.06NQIEE404 pKa = 4.17FLGIQIDD411 pKa = 3.82KK412 pKa = 11.11NGIKK416 pKa = 8.84MQTHH420 pKa = 6.44IVQKK424 pKa = 10.46IINLDD429 pKa = 3.6EE430 pKa = 5.32NIDD433 pKa = 3.95TKK435 pKa = 11.26KK436 pKa = 10.64KK437 pKa = 9.4LQSFLGIVNQVRR449 pKa = 11.84EE450 pKa = 4.78YY451 pKa = 10.52IPKK454 pKa = 9.98LAEE457 pKa = 3.88NLKK460 pKa = 10.03PLQKK464 pKa = 10.38KK465 pKa = 9.34LKK467 pKa = 10.02KK468 pKa = 10.1DD469 pKa = 3.28VEE471 pKa = 4.3YY472 pKa = 11.37SFDD475 pKa = 3.92EE476 pKa = 4.36KK477 pKa = 11.22DD478 pKa = 3.05KK479 pKa = 11.27EE480 pKa = 4.13QIKK483 pKa = 10.16KK484 pKa = 10.37IKK486 pKa = 9.72ILCKK490 pKa = 10.23KK491 pKa = 10.06LPKK494 pKa = 10.31LYY496 pKa = 10.72FPDD499 pKa = 3.63EE500 pKa = 4.39NKK502 pKa = 10.17KK503 pKa = 8.68FTYY506 pKa = 9.92IVEE509 pKa = 4.44TDD511 pKa = 3.28SSNYY515 pKa = 10.37SYY517 pKa = 11.63GGVLKK522 pKa = 10.7YY523 pKa = 10.35RR524 pKa = 11.84YY525 pKa = 8.69NKK527 pKa = 9.98EE528 pKa = 4.13KK529 pKa = 10.28IEE531 pKa = 3.99HH532 pKa = 5.78HH533 pKa = 6.1CRR535 pKa = 11.84YY536 pKa = 10.05YY537 pKa = 10.68SGSYY541 pKa = 9.4TEE543 pKa = 4.76PQEE546 pKa = 3.65KK547 pKa = 9.45WEE549 pKa = 4.18INRR552 pKa = 11.84KK553 pKa = 8.0EE554 pKa = 4.06LFALYY559 pKa = 10.07KK560 pKa = 10.4CLLAFEE566 pKa = 4.93PYY568 pKa = 9.5IVYY571 pKa = 9.05TRR573 pKa = 11.84FIVRR577 pKa = 11.84TDD579 pKa = 3.16NTQVKK584 pKa = 7.71WWITRR589 pKa = 11.84KK590 pKa = 9.67VQDD593 pKa = 3.37SVTTKK598 pKa = 10.13EE599 pKa = 3.62IRR601 pKa = 11.84RR602 pKa = 11.84LVLNILNFTFTIEE615 pKa = 4.96IINTNKK621 pKa = 10.13NVVADD626 pKa = 3.77YY627 pKa = 10.86LSRR630 pKa = 11.84QSYY633 pKa = 9.84PNN635 pKa = 3.3
Molecular weight: 75.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2050 |
417 |
635 |
512.5 |
60.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.024 ± 0.387 | 1.463 ± 0.209 |
4.439 ± 0.499 | 9.902 ± 1.288 |
2.927 ± 0.535 | 3.073 ± 0.21 |
1.561 ± 0.298 | 11.61 ± 1.333 |
12.146 ± 1.178 | 7.659 ± 0.413 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.805 ± 0.272 | 7.317 ± 0.347 |
3.366 ± 0.313 | 4.39 ± 0.437 |
3.805 ± 0.231 | 4.683 ± 0.439 |
7.024 ± 0.486 | 3.073 ± 0.686 |
1.073 ± 0.223 | 5.659 ± 0.26 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
