
Puma lentivirus 14
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus; Puma lentivirus
Average proteome isoelectric point is 7.17
Get precalculated fractions of proteins
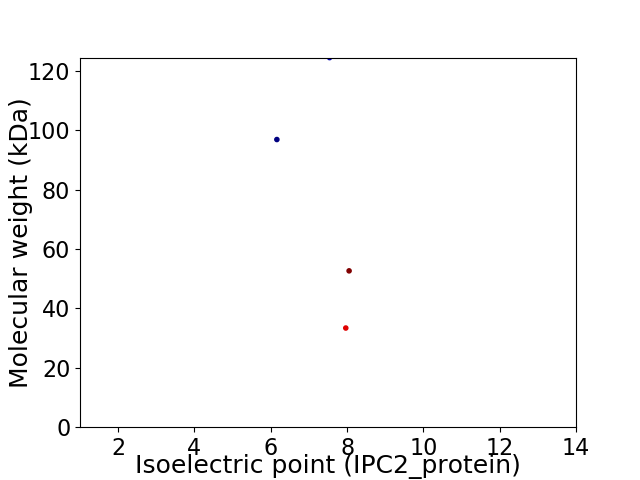
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q84811|Q84811_9RETR Env polyprotein OS=Puma lentivirus 14 OX=32615 GN=env PE=4 SV=1
MM1 pKa = 7.19AQANPMDD8 pKa = 4.41NAWIGPEE15 pKa = 3.97EE16 pKa = 4.98GEE18 pKa = 4.08MLLDD22 pKa = 3.84FEE24 pKa = 5.12VVMMLHH30 pKa = 6.73EE31 pKa = 4.72GSNYY35 pKa = 9.82PGINPFRR42 pKa = 11.84DD43 pKa = 3.47PTLTPEE49 pKa = 4.72KK50 pKa = 9.89KK51 pKa = 9.7QQTAALLQPLLVRR64 pKa = 11.84IKK66 pKa = 10.87QNLLQQGKK74 pKa = 8.61VKK76 pKa = 10.56NWGKK80 pKa = 10.3QDD82 pKa = 3.11WDD84 pKa = 2.91KK85 pKa = 10.79MRR87 pKa = 11.84RR88 pKa = 11.84YY89 pKa = 9.73KK90 pKa = 10.71AGEE93 pKa = 3.83RR94 pKa = 11.84AWIKK98 pKa = 9.68FVKK101 pKa = 10.19IFGGIHH107 pKa = 5.85NMPKK111 pKa = 10.43EE112 pKa = 3.99EE113 pKa = 4.43DD114 pKa = 3.44LGTFSGDD121 pKa = 2.43IDD123 pKa = 4.4MITPHH128 pKa = 7.02SYY130 pKa = 10.83RR131 pKa = 11.84KK132 pKa = 8.29VTYY135 pKa = 8.49NWKK138 pKa = 8.86TSLCFFLLIGSAMLAYY154 pKa = 10.56SLIFTIIIKK163 pKa = 10.01QALAQAVTLALDD175 pKa = 4.15PPWVIPLKK183 pKa = 10.47YY184 pKa = 10.15RR185 pKa = 11.84EE186 pKa = 5.18DD187 pKa = 3.6INFQCIGNHH196 pKa = 5.85PEE198 pKa = 3.93CNLPKK203 pKa = 10.54EE204 pKa = 4.28VGDD207 pKa = 3.47WKK209 pKa = 11.37QNFTWVYY216 pKa = 5.99QTPINEE222 pKa = 4.54TIGLEE227 pKa = 4.03IYY229 pKa = 10.11AQEE232 pKa = 3.98IAAKK236 pKa = 9.97LWQQVFLQCKK246 pKa = 9.58KK247 pKa = 10.73GPRR250 pKa = 11.84DD251 pKa = 3.41LDD253 pKa = 3.56TAIKK257 pKa = 10.35YY258 pKa = 7.42WICFYY263 pKa = 10.58DD264 pKa = 3.57TAIKK268 pKa = 10.43YY269 pKa = 10.24LLGYY273 pKa = 8.01EE274 pKa = 4.92HH275 pKa = 7.21IQLCPLGGYY284 pKa = 9.47LVYY287 pKa = 11.01DD288 pKa = 3.69NTTKK292 pKa = 10.34EE293 pKa = 4.09ISMCTPPLSLRR304 pKa = 11.84LLNFTLSQEE313 pKa = 3.95KK314 pKa = 9.31WEE316 pKa = 4.3QEE318 pKa = 4.0PFTDD322 pKa = 5.04IVWFGNKK329 pKa = 9.38ALNTTVNNITQVQINVTMVCNVIVPEE355 pKa = 3.96QVGKK359 pKa = 10.77KK360 pKa = 9.57KK361 pKa = 10.71GRR363 pKa = 11.84FQFYY367 pKa = 11.08NEE369 pKa = 4.38FLGPWGGGRR378 pKa = 11.84FQEE381 pKa = 4.86IIVRR385 pKa = 11.84YY386 pKa = 8.38QDD388 pKa = 2.98WVNISDD394 pKa = 5.54PILDD398 pKa = 4.64LNCSGIPGVDD408 pKa = 4.1FNHH411 pKa = 6.73TEE413 pKa = 3.82GNYY416 pKa = 9.13TCIHH420 pKa = 6.26NKK422 pKa = 9.01TYY424 pKa = 10.49QEE426 pKa = 4.29GDD428 pKa = 2.81ICTQPEE434 pKa = 5.06FIAPCYY440 pKa = 9.2NSNYY444 pKa = 9.85SIPLVVHH451 pKa = 6.52CKK453 pKa = 9.76LHH455 pKa = 6.05NEE457 pKa = 4.39NITGTVLQVMRR468 pKa = 11.84CRR470 pKa = 11.84GMKK473 pKa = 10.47DD474 pKa = 2.83LDD476 pKa = 3.58LRR478 pKa = 11.84IAGEE482 pKa = 4.28FVTLNLTLVKK492 pKa = 10.68DD493 pKa = 3.67PFLDD497 pKa = 3.58YY498 pKa = 11.17LRR500 pKa = 11.84NQVNYY505 pKa = 9.06TCTLNGTFWVYY516 pKa = 11.03KK517 pKa = 9.41SNKK520 pKa = 8.68PKK522 pKa = 9.91WDD524 pKa = 3.74TNEE527 pKa = 3.88TIYY530 pKa = 11.33APVSNFTDD538 pKa = 3.69DD539 pKa = 3.4RR540 pKa = 11.84VVWGAYY546 pKa = 9.96NSILYY551 pKa = 9.55QFYY554 pKa = 11.03ALQKK558 pKa = 10.49FKK560 pKa = 11.07LLKK563 pKa = 10.57KK564 pKa = 9.75PVATIMPPVRR574 pKa = 11.84EE575 pKa = 3.79PHH577 pKa = 6.09RR578 pKa = 11.84RR579 pKa = 11.84QTRR582 pKa = 11.84EE583 pKa = 3.57LRR585 pKa = 11.84QKK587 pKa = 10.53RR588 pKa = 11.84GLGLTIAIVGAVTAGMIGTTTGTAALAVSVRR619 pKa = 11.84LKK621 pKa = 10.99EE622 pKa = 4.36VMLQQAHH629 pKa = 6.72INEE632 pKa = 4.36QVLGALRR639 pKa = 11.84IVQRR643 pKa = 11.84RR644 pKa = 11.84LQDD647 pKa = 3.12AEE649 pKa = 4.27RR650 pKa = 11.84FILSLHH656 pKa = 5.11QRR658 pKa = 11.84VTKK661 pKa = 10.23IEE663 pKa = 4.11RR664 pKa = 11.84FLEE667 pKa = 3.85IQYY670 pKa = 10.05QLQGMCPFKK679 pKa = 10.63DD680 pKa = 3.05ICQLDD685 pKa = 3.66MNFNFTDD692 pKa = 4.3YY693 pKa = 11.23NDD695 pKa = 3.53SWPMGRR701 pKa = 11.84WAEE704 pKa = 3.99QAEE707 pKa = 4.33KK708 pKa = 10.5DD709 pKa = 3.27WRR711 pKa = 11.84EE712 pKa = 3.88FQSLIDD718 pKa = 3.42NATRR722 pKa = 11.84SNEE725 pKa = 3.88NLKK728 pKa = 9.49TDD730 pKa = 3.71LTKK733 pKa = 11.04LEE735 pKa = 4.06VLDD738 pKa = 3.69WFSWFPINSVFSTIFSLIIIIIVVVLARR766 pKa = 11.84PCLEE770 pKa = 3.75NCIKK774 pKa = 10.94GFFSMLKK781 pKa = 9.19GYY783 pKa = 10.45RR784 pKa = 11.84PIKK787 pKa = 9.71LQMVEE792 pKa = 4.25IPLEE796 pKa = 4.25GTTQEE801 pKa = 4.27EE802 pKa = 4.36EE803 pKa = 4.51GEE805 pKa = 4.25EE806 pKa = 4.14EE807 pKa = 4.19DD808 pKa = 5.21NEE810 pKa = 4.85DD811 pKa = 4.69GGEE814 pKa = 4.17SCQTWRR820 pKa = 11.84SDD822 pKa = 4.75LNNCLGKK829 pKa = 10.51KK830 pKa = 10.04KK831 pKa = 10.38SLKK834 pKa = 9.31GLNMHH839 pKa = 7.15HH840 pKa = 7.0
MM1 pKa = 7.19AQANPMDD8 pKa = 4.41NAWIGPEE15 pKa = 3.97EE16 pKa = 4.98GEE18 pKa = 4.08MLLDD22 pKa = 3.84FEE24 pKa = 5.12VVMMLHH30 pKa = 6.73EE31 pKa = 4.72GSNYY35 pKa = 9.82PGINPFRR42 pKa = 11.84DD43 pKa = 3.47PTLTPEE49 pKa = 4.72KK50 pKa = 9.89KK51 pKa = 9.7QQTAALLQPLLVRR64 pKa = 11.84IKK66 pKa = 10.87QNLLQQGKK74 pKa = 8.61VKK76 pKa = 10.56NWGKK80 pKa = 10.3QDD82 pKa = 3.11WDD84 pKa = 2.91KK85 pKa = 10.79MRR87 pKa = 11.84RR88 pKa = 11.84YY89 pKa = 9.73KK90 pKa = 10.71AGEE93 pKa = 3.83RR94 pKa = 11.84AWIKK98 pKa = 9.68FVKK101 pKa = 10.19IFGGIHH107 pKa = 5.85NMPKK111 pKa = 10.43EE112 pKa = 3.99EE113 pKa = 4.43DD114 pKa = 3.44LGTFSGDD121 pKa = 2.43IDD123 pKa = 4.4MITPHH128 pKa = 7.02SYY130 pKa = 10.83RR131 pKa = 11.84KK132 pKa = 8.29VTYY135 pKa = 8.49NWKK138 pKa = 8.86TSLCFFLLIGSAMLAYY154 pKa = 10.56SLIFTIIIKK163 pKa = 10.01QALAQAVTLALDD175 pKa = 4.15PPWVIPLKK183 pKa = 10.47YY184 pKa = 10.15RR185 pKa = 11.84EE186 pKa = 5.18DD187 pKa = 3.6INFQCIGNHH196 pKa = 5.85PEE198 pKa = 3.93CNLPKK203 pKa = 10.54EE204 pKa = 4.28VGDD207 pKa = 3.47WKK209 pKa = 11.37QNFTWVYY216 pKa = 5.99QTPINEE222 pKa = 4.54TIGLEE227 pKa = 4.03IYY229 pKa = 10.11AQEE232 pKa = 3.98IAAKK236 pKa = 9.97LWQQVFLQCKK246 pKa = 9.58KK247 pKa = 10.73GPRR250 pKa = 11.84DD251 pKa = 3.41LDD253 pKa = 3.56TAIKK257 pKa = 10.35YY258 pKa = 7.42WICFYY263 pKa = 10.58DD264 pKa = 3.57TAIKK268 pKa = 10.43YY269 pKa = 10.24LLGYY273 pKa = 8.01EE274 pKa = 4.92HH275 pKa = 7.21IQLCPLGGYY284 pKa = 9.47LVYY287 pKa = 11.01DD288 pKa = 3.69NTTKK292 pKa = 10.34EE293 pKa = 4.09ISMCTPPLSLRR304 pKa = 11.84LLNFTLSQEE313 pKa = 3.95KK314 pKa = 9.31WEE316 pKa = 4.3QEE318 pKa = 4.0PFTDD322 pKa = 5.04IVWFGNKK329 pKa = 9.38ALNTTVNNITQVQINVTMVCNVIVPEE355 pKa = 3.96QVGKK359 pKa = 10.77KK360 pKa = 9.57KK361 pKa = 10.71GRR363 pKa = 11.84FQFYY367 pKa = 11.08NEE369 pKa = 4.38FLGPWGGGRR378 pKa = 11.84FQEE381 pKa = 4.86IIVRR385 pKa = 11.84YY386 pKa = 8.38QDD388 pKa = 2.98WVNISDD394 pKa = 5.54PILDD398 pKa = 4.64LNCSGIPGVDD408 pKa = 4.1FNHH411 pKa = 6.73TEE413 pKa = 3.82GNYY416 pKa = 9.13TCIHH420 pKa = 6.26NKK422 pKa = 9.01TYY424 pKa = 10.49QEE426 pKa = 4.29GDD428 pKa = 2.81ICTQPEE434 pKa = 5.06FIAPCYY440 pKa = 9.2NSNYY444 pKa = 9.85SIPLVVHH451 pKa = 6.52CKK453 pKa = 9.76LHH455 pKa = 6.05NEE457 pKa = 4.39NITGTVLQVMRR468 pKa = 11.84CRR470 pKa = 11.84GMKK473 pKa = 10.47DD474 pKa = 2.83LDD476 pKa = 3.58LRR478 pKa = 11.84IAGEE482 pKa = 4.28FVTLNLTLVKK492 pKa = 10.68DD493 pKa = 3.67PFLDD497 pKa = 3.58YY498 pKa = 11.17LRR500 pKa = 11.84NQVNYY505 pKa = 9.06TCTLNGTFWVYY516 pKa = 11.03KK517 pKa = 9.41SNKK520 pKa = 8.68PKK522 pKa = 9.91WDD524 pKa = 3.74TNEE527 pKa = 3.88TIYY530 pKa = 11.33APVSNFTDD538 pKa = 3.69DD539 pKa = 3.4RR540 pKa = 11.84VVWGAYY546 pKa = 9.96NSILYY551 pKa = 9.55QFYY554 pKa = 11.03ALQKK558 pKa = 10.49FKK560 pKa = 11.07LLKK563 pKa = 10.57KK564 pKa = 9.75PVATIMPPVRR574 pKa = 11.84EE575 pKa = 3.79PHH577 pKa = 6.09RR578 pKa = 11.84RR579 pKa = 11.84QTRR582 pKa = 11.84EE583 pKa = 3.57LRR585 pKa = 11.84QKK587 pKa = 10.53RR588 pKa = 11.84GLGLTIAIVGAVTAGMIGTTTGTAALAVSVRR619 pKa = 11.84LKK621 pKa = 10.99EE622 pKa = 4.36VMLQQAHH629 pKa = 6.72INEE632 pKa = 4.36QVLGALRR639 pKa = 11.84IVQRR643 pKa = 11.84RR644 pKa = 11.84LQDD647 pKa = 3.12AEE649 pKa = 4.27RR650 pKa = 11.84FILSLHH656 pKa = 5.11QRR658 pKa = 11.84VTKK661 pKa = 10.23IEE663 pKa = 4.11RR664 pKa = 11.84FLEE667 pKa = 3.85IQYY670 pKa = 10.05QLQGMCPFKK679 pKa = 10.63DD680 pKa = 3.05ICQLDD685 pKa = 3.66MNFNFTDD692 pKa = 4.3YY693 pKa = 11.23NDD695 pKa = 3.53SWPMGRR701 pKa = 11.84WAEE704 pKa = 3.99QAEE707 pKa = 4.33KK708 pKa = 10.5DD709 pKa = 3.27WRR711 pKa = 11.84EE712 pKa = 3.88FQSLIDD718 pKa = 3.42NATRR722 pKa = 11.84SNEE725 pKa = 3.88NLKK728 pKa = 9.49TDD730 pKa = 3.71LTKK733 pKa = 11.04LEE735 pKa = 4.06VLDD738 pKa = 3.69WFSWFPINSVFSTIFSLIIIIIVVVLARR766 pKa = 11.84PCLEE770 pKa = 3.75NCIKK774 pKa = 10.94GFFSMLKK781 pKa = 9.19GYY783 pKa = 10.45RR784 pKa = 11.84PIKK787 pKa = 9.71LQMVEE792 pKa = 4.25IPLEE796 pKa = 4.25GTTQEE801 pKa = 4.27EE802 pKa = 4.36EE803 pKa = 4.51GEE805 pKa = 4.25EE806 pKa = 4.14EE807 pKa = 4.19DD808 pKa = 5.21NEE810 pKa = 4.85DD811 pKa = 4.69GGEE814 pKa = 4.17SCQTWRR820 pKa = 11.84SDD822 pKa = 4.75LNNCLGKK829 pKa = 10.51KK830 pKa = 10.04KK831 pKa = 10.38SLKK834 pKa = 9.31GLNMHH839 pKa = 7.15HH840 pKa = 7.0
Molecular weight: 96.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84809|Q84809_9RETR Exoribonuclease H OS=Puma lentivirus 14 OX=32615 GN=pol PE=3 SV=1
MM1 pKa = 7.66GNNQGKK7 pKa = 7.71EE8 pKa = 3.71LKK10 pKa = 10.24AALRR14 pKa = 11.84RR15 pKa = 11.84ACNVTVGEE23 pKa = 4.51GKK25 pKa = 9.85RR26 pKa = 11.84SKK28 pKa = 10.66RR29 pKa = 11.84YY30 pKa = 8.34TEE32 pKa = 4.49GNLMWAIKK40 pKa = 10.0FGNACTGRR48 pKa = 11.84DD49 pKa = 3.47PADD52 pKa = 3.38VPEE55 pKa = 4.2TLVEE59 pKa = 3.76IRR61 pKa = 11.84NFIHH65 pKa = 6.9EE66 pKa = 4.75LQDD69 pKa = 3.32KK70 pKa = 8.85LQKK73 pKa = 10.68FGGSKK78 pKa = 9.83EE79 pKa = 4.01LEE81 pKa = 4.1NCIKK85 pKa = 9.02TLKK88 pKa = 9.79VLTVAGVLKK97 pKa = 10.26LPCQNTEE104 pKa = 4.07SAIKK108 pKa = 10.31LYY110 pKa = 9.72EE111 pKa = 4.09TMGLLGPATDD121 pKa = 4.96KK122 pKa = 11.15KK123 pKa = 10.69IEE125 pKa = 4.17EE126 pKa = 4.19NLEE129 pKa = 3.98EE130 pKa = 4.87KK131 pKa = 10.15PAEE134 pKa = 4.33AYY136 pKa = 8.52PVQVANGVHH145 pKa = 4.64QHH147 pKa = 5.2VSFNPRR153 pKa = 11.84TAAIWMEE160 pKa = 4.15KK161 pKa = 10.27ARR163 pKa = 11.84GGLGSEE169 pKa = 4.42EE170 pKa = 3.72AVLWFTAFSADD181 pKa = 3.66LTATDD186 pKa = 3.87MASLITAAPGCAADD200 pKa = 5.58KK201 pKa = 10.91KK202 pKa = 10.98IIDD205 pKa = 4.58DD206 pKa = 3.83KK207 pKa = 11.52LKK209 pKa = 10.91EE210 pKa = 4.06LTAKK214 pKa = 10.1YY215 pKa = 10.3AQDD218 pKa = 4.0HH219 pKa = 6.95PDD221 pKa = 3.33GPRR224 pKa = 11.84PLPYY228 pKa = 9.33FTAEE232 pKa = 4.08EE233 pKa = 4.05IMGIGIPQNVQSQPQYY249 pKa = 11.06GPARR253 pKa = 11.84AQARR257 pKa = 11.84LWFLEE262 pKa = 4.09ALGHH266 pKa = 5.05LQKK269 pKa = 10.66IKK271 pKa = 10.69AGEE274 pKa = 4.04PKK276 pKa = 10.45AVTLRR281 pKa = 11.84QGPKK285 pKa = 9.74EE286 pKa = 4.17SYY288 pKa = 10.61KK289 pKa = 11.13DD290 pKa = 3.76FIDD293 pKa = 5.28RR294 pKa = 11.84LFQQIDD300 pKa = 3.33QEE302 pKa = 4.38QASDD306 pKa = 3.68EE307 pKa = 4.09VRR309 pKa = 11.84DD310 pKa = 3.83YY311 pKa = 11.58LKK313 pKa = 10.78QSLSISNANGEE324 pKa = 4.26CRR326 pKa = 11.84KK327 pKa = 10.49AMTHH331 pKa = 6.37LRR333 pKa = 11.84PEE335 pKa = 4.11STLEE339 pKa = 3.8EE340 pKa = 4.32KK341 pKa = 10.73LRR343 pKa = 11.84ACQDD347 pKa = 2.99IGSTQYY353 pKa = 11.27KK354 pKa = 8.14MQMLAEE360 pKa = 5.15AFNQMQVNQVQRR372 pKa = 11.84GGFRR376 pKa = 11.84GGRR379 pKa = 11.84GGNRR383 pKa = 11.84GRR385 pKa = 11.84GGRR388 pKa = 11.84GRR390 pKa = 11.84GRR392 pKa = 11.84GRR394 pKa = 11.84GLGPLNCFNCGKK406 pKa = 9.15PGHH409 pKa = 6.94LASQCRR415 pKa = 11.84QPIKK419 pKa = 10.3CYY421 pKa = 9.11KK422 pKa = 10.09CGGSGHH428 pKa = 7.11LAIDD432 pKa = 3.78CLGGNDD438 pKa = 4.42SKK440 pKa = 11.51NGQNRR445 pKa = 11.84GTAAPRR451 pKa = 11.84QFQVQQNNTLYY462 pKa = 10.19PSLKK466 pKa = 10.26EE467 pKa = 3.97MQTEE471 pKa = 4.23PTAPPMEE478 pKa = 4.27II479 pKa = 3.82
MM1 pKa = 7.66GNNQGKK7 pKa = 7.71EE8 pKa = 3.71LKK10 pKa = 10.24AALRR14 pKa = 11.84RR15 pKa = 11.84ACNVTVGEE23 pKa = 4.51GKK25 pKa = 9.85RR26 pKa = 11.84SKK28 pKa = 10.66RR29 pKa = 11.84YY30 pKa = 8.34TEE32 pKa = 4.49GNLMWAIKK40 pKa = 10.0FGNACTGRR48 pKa = 11.84DD49 pKa = 3.47PADD52 pKa = 3.38VPEE55 pKa = 4.2TLVEE59 pKa = 3.76IRR61 pKa = 11.84NFIHH65 pKa = 6.9EE66 pKa = 4.75LQDD69 pKa = 3.32KK70 pKa = 8.85LQKK73 pKa = 10.68FGGSKK78 pKa = 9.83EE79 pKa = 4.01LEE81 pKa = 4.1NCIKK85 pKa = 9.02TLKK88 pKa = 9.79VLTVAGVLKK97 pKa = 10.26LPCQNTEE104 pKa = 4.07SAIKK108 pKa = 10.31LYY110 pKa = 9.72EE111 pKa = 4.09TMGLLGPATDD121 pKa = 4.96KK122 pKa = 11.15KK123 pKa = 10.69IEE125 pKa = 4.17EE126 pKa = 4.19NLEE129 pKa = 3.98EE130 pKa = 4.87KK131 pKa = 10.15PAEE134 pKa = 4.33AYY136 pKa = 8.52PVQVANGVHH145 pKa = 4.64QHH147 pKa = 5.2VSFNPRR153 pKa = 11.84TAAIWMEE160 pKa = 4.15KK161 pKa = 10.27ARR163 pKa = 11.84GGLGSEE169 pKa = 4.42EE170 pKa = 3.72AVLWFTAFSADD181 pKa = 3.66LTATDD186 pKa = 3.87MASLITAAPGCAADD200 pKa = 5.58KK201 pKa = 10.91KK202 pKa = 10.98IIDD205 pKa = 4.58DD206 pKa = 3.83KK207 pKa = 11.52LKK209 pKa = 10.91EE210 pKa = 4.06LTAKK214 pKa = 10.1YY215 pKa = 10.3AQDD218 pKa = 4.0HH219 pKa = 6.95PDD221 pKa = 3.33GPRR224 pKa = 11.84PLPYY228 pKa = 9.33FTAEE232 pKa = 4.08EE233 pKa = 4.05IMGIGIPQNVQSQPQYY249 pKa = 11.06GPARR253 pKa = 11.84AQARR257 pKa = 11.84LWFLEE262 pKa = 4.09ALGHH266 pKa = 5.05LQKK269 pKa = 10.66IKK271 pKa = 10.69AGEE274 pKa = 4.04PKK276 pKa = 10.45AVTLRR281 pKa = 11.84QGPKK285 pKa = 9.74EE286 pKa = 4.17SYY288 pKa = 10.61KK289 pKa = 11.13DD290 pKa = 3.76FIDD293 pKa = 5.28RR294 pKa = 11.84LFQQIDD300 pKa = 3.33QEE302 pKa = 4.38QASDD306 pKa = 3.68EE307 pKa = 4.09VRR309 pKa = 11.84DD310 pKa = 3.83YY311 pKa = 11.58LKK313 pKa = 10.78QSLSISNANGEE324 pKa = 4.26CRR326 pKa = 11.84KK327 pKa = 10.49AMTHH331 pKa = 6.37LRR333 pKa = 11.84PEE335 pKa = 4.11STLEE339 pKa = 3.8EE340 pKa = 4.32KK341 pKa = 10.73LRR343 pKa = 11.84ACQDD347 pKa = 2.99IGSTQYY353 pKa = 11.27KK354 pKa = 8.14MQMLAEE360 pKa = 5.15AFNQMQVNQVQRR372 pKa = 11.84GGFRR376 pKa = 11.84GGRR379 pKa = 11.84GGNRR383 pKa = 11.84GRR385 pKa = 11.84GGRR388 pKa = 11.84GRR390 pKa = 11.84GRR392 pKa = 11.84GRR394 pKa = 11.84GLGPLNCFNCGKK406 pKa = 9.15PGHH409 pKa = 6.94LASQCRR415 pKa = 11.84QPIKK419 pKa = 10.3CYY421 pKa = 9.11KK422 pKa = 10.09CGGSGHH428 pKa = 7.11LAIDD432 pKa = 3.78CLGGNDD438 pKa = 4.42SKK440 pKa = 11.51NGQNRR445 pKa = 11.84GTAAPRR451 pKa = 11.84QFQVQQNNTLYY462 pKa = 10.19PSLKK466 pKa = 10.26EE467 pKa = 3.97MQTEE471 pKa = 4.23PTAPPMEE478 pKa = 4.27II479 pKa = 3.82
Molecular weight: 52.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2681 |
276 |
1086 |
670.3 |
76.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.147 ± 0.983 | 2.201 ± 0.462 |
4.998 ± 0.352 | 6.527 ± 0.433 |
3.394 ± 0.495 | 7.162 ± 0.815 |
1.641 ± 0.097 | 7.535 ± 1.079 |
7.46 ± 0.574 | 9.549 ± 0.404 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.536 ± 0.133 | 5.259 ± 0.38 |
5.035 ± 0.149 | 6.117 ± 0.247 |
4.924 ± 0.546 | 3.916 ± 0.196 |
5.073 ± 0.551 | 5.334 ± 0.532 |
2.648 ± 0.687 | 3.543 ± 0.377 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
