
Pelodiscus sinensis (Chinese softshell turtle) (Trionyx sinensis)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida;
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
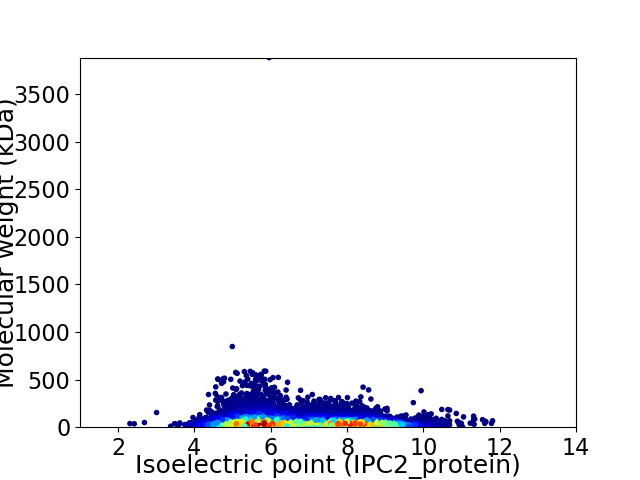
Virtual 2D-PAGE plot for 20509 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K7FRB3|K7FRB3_PELSI Solute carrier family 11 member 2 OS=Pelodiscus sinensis OX=13735 GN=SLC11A2 PE=3 SV=1
MM1 pKa = 8.37DD2 pKa = 5.04DD3 pKa = 3.3TTNNQFSYY11 pKa = 9.39VTHH14 pKa = 7.13EE15 pKa = 5.05DD16 pKa = 2.66ICNCFNGDD24 pKa = 3.43TLLAIQAPCGTQLEE38 pKa = 4.63VPIPEE43 pKa = 4.4MGQNGQKK50 pKa = 10.21KK51 pKa = 8.37YY52 pKa = 10.6QISLKK57 pKa = 10.33SSSGPIQVLLINKK70 pKa = 7.64EE71 pKa = 4.42SSSSTPVVFPVPPPDD86 pKa = 6.27DD87 pKa = 3.32IAQPPSQLATPVTPPKK103 pKa = 9.34PTATTEE109 pKa = 4.13NTSEE113 pKa = 3.84QHH115 pKa = 6.05SVNEE119 pKa = 4.35GQHH122 pKa = 5.97LPRR125 pKa = 11.84TTDD128 pKa = 3.24TPSDD132 pKa = 3.23NSAQEE137 pKa = 3.91NTTPEE142 pKa = 4.2TPYY145 pKa = 11.53SNLPDD150 pKa = 3.53SLLYY154 pKa = 7.89TTLATDD160 pKa = 4.12TTQTTVNSNDD170 pKa = 3.51YY171 pKa = 10.43QALLPLDD178 pKa = 4.05VNCILKK184 pKa = 10.4PNSLDD189 pKa = 3.35MAKK192 pKa = 9.27MDD194 pKa = 4.58EE195 pKa = 4.24PTGTISSDD203 pKa = 3.37IIDD206 pKa = 4.36EE207 pKa = 4.39LMSSDD212 pKa = 3.97VFPLLRR218 pKa = 11.84LSPTPGDD225 pKa = 3.86DD226 pKa = 3.93YY227 pKa = 11.79NFNLDD232 pKa = 4.1DD233 pKa = 4.45SEE235 pKa = 4.88GVCDD239 pKa = 5.38LFDD242 pKa = 3.89VQILNYY248 pKa = 10.41
MM1 pKa = 8.37DD2 pKa = 5.04DD3 pKa = 3.3TTNNQFSYY11 pKa = 9.39VTHH14 pKa = 7.13EE15 pKa = 5.05DD16 pKa = 2.66ICNCFNGDD24 pKa = 3.43TLLAIQAPCGTQLEE38 pKa = 4.63VPIPEE43 pKa = 4.4MGQNGQKK50 pKa = 10.21KK51 pKa = 8.37YY52 pKa = 10.6QISLKK57 pKa = 10.33SSSGPIQVLLINKK70 pKa = 7.64EE71 pKa = 4.42SSSSTPVVFPVPPPDD86 pKa = 6.27DD87 pKa = 3.32IAQPPSQLATPVTPPKK103 pKa = 9.34PTATTEE109 pKa = 4.13NTSEE113 pKa = 3.84QHH115 pKa = 6.05SVNEE119 pKa = 4.35GQHH122 pKa = 5.97LPRR125 pKa = 11.84TTDD128 pKa = 3.24TPSDD132 pKa = 3.23NSAQEE137 pKa = 3.91NTTPEE142 pKa = 4.2TPYY145 pKa = 11.53SNLPDD150 pKa = 3.53SLLYY154 pKa = 7.89TTLATDD160 pKa = 4.12TTQTTVNSNDD170 pKa = 3.51YY171 pKa = 10.43QALLPLDD178 pKa = 4.05VNCILKK184 pKa = 10.4PNSLDD189 pKa = 3.35MAKK192 pKa = 9.27MDD194 pKa = 4.58EE195 pKa = 4.24PTGTISSDD203 pKa = 3.37IIDD206 pKa = 4.36EE207 pKa = 4.39LMSSDD212 pKa = 3.97VFPLLRR218 pKa = 11.84LSPTPGDD225 pKa = 3.86DD226 pKa = 3.93YY227 pKa = 11.79NFNLDD232 pKa = 4.1DD233 pKa = 4.45SEE235 pKa = 4.88GVCDD239 pKa = 5.38LFDD242 pKa = 3.89VQILNYY248 pKa = 10.41
Molecular weight: 26.97 kDa
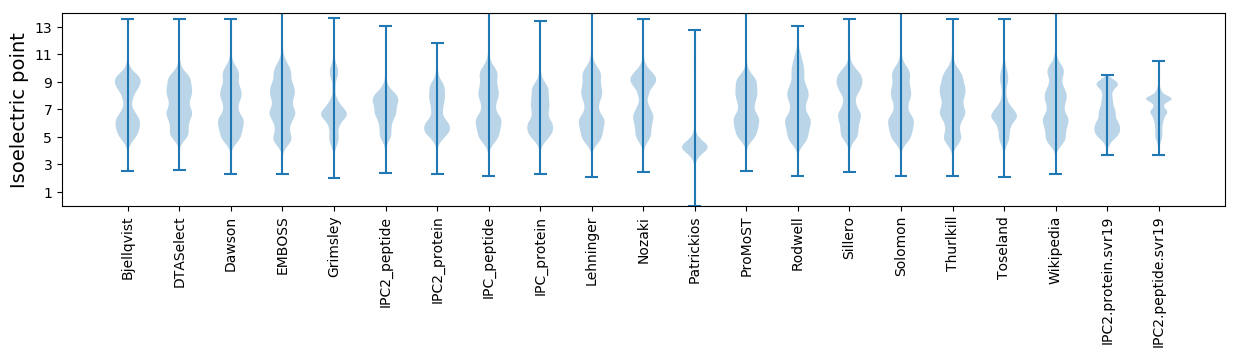
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7F0I4|K7F0I4_PELSI Reverse transcriptase domain-containing protein OS=Pelodiscus sinensis OX=13735 PE=4 SV=1
MM1 pKa = 7.66PGSPARR7 pKa = 11.84PGQWGLVGGAGGLGGRR23 pKa = 11.84MPGSPARR30 pKa = 11.84PGQWGLVGGAGGLGGRR46 pKa = 11.84MPGSPARR53 pKa = 11.84PGQWGLVGGAGGLGGRR69 pKa = 11.84MPGSPARR76 pKa = 11.84PGQWGLVGGAGGLGGRR92 pKa = 11.84MPGSPARR99 pKa = 11.84PGQWGLVGGAGGLGGRR115 pKa = 11.84MPGSPARR122 pKa = 11.84PGQWGLVGGAGGLGGRR138 pKa = 11.84MPGSPARR145 pKa = 11.84PGQWGLVGGAGGLGGRR161 pKa = 11.84MPGSPARR168 pKa = 11.84PGQWGLVGGAGGLGGRR184 pKa = 11.84MPGSPARR191 pKa = 11.84PGQWGLVGGAGGLGGRR207 pKa = 11.84MPGSPARR214 pKa = 11.84PGQWGLVGGAGGLGGRR230 pKa = 11.84MPGSPARR237 pKa = 11.84PGQWGLVGGAGGLGGRR253 pKa = 11.84MPGSPARR260 pKa = 11.84PGQWGLVGGAGGLGGRR276 pKa = 11.84MPGSPARR283 pKa = 11.84PGQGAGQGCGAGPRR297 pKa = 11.84LHH299 pKa = 6.87GGGAVSHH306 pKa = 6.26LRR308 pKa = 11.84GAGSPHH314 pKa = 5.63QGVWASFSSRR324 pKa = 11.84SSS326 pKa = 3.04
MM1 pKa = 7.66PGSPARR7 pKa = 11.84PGQWGLVGGAGGLGGRR23 pKa = 11.84MPGSPARR30 pKa = 11.84PGQWGLVGGAGGLGGRR46 pKa = 11.84MPGSPARR53 pKa = 11.84PGQWGLVGGAGGLGGRR69 pKa = 11.84MPGSPARR76 pKa = 11.84PGQWGLVGGAGGLGGRR92 pKa = 11.84MPGSPARR99 pKa = 11.84PGQWGLVGGAGGLGGRR115 pKa = 11.84MPGSPARR122 pKa = 11.84PGQWGLVGGAGGLGGRR138 pKa = 11.84MPGSPARR145 pKa = 11.84PGQWGLVGGAGGLGGRR161 pKa = 11.84MPGSPARR168 pKa = 11.84PGQWGLVGGAGGLGGRR184 pKa = 11.84MPGSPARR191 pKa = 11.84PGQWGLVGGAGGLGGRR207 pKa = 11.84MPGSPARR214 pKa = 11.84PGQWGLVGGAGGLGGRR230 pKa = 11.84MPGSPARR237 pKa = 11.84PGQWGLVGGAGGLGGRR253 pKa = 11.84MPGSPARR260 pKa = 11.84PGQWGLVGGAGGLGGRR276 pKa = 11.84MPGSPARR283 pKa = 11.84PGQGAGQGCGAGPRR297 pKa = 11.84LHH299 pKa = 6.87GGGAVSHH306 pKa = 6.26LRR308 pKa = 11.84GAGSPHH314 pKa = 5.63QGVWASFSSRR324 pKa = 11.84SSS326 pKa = 3.04
Molecular weight: 30.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
10850553 |
20 |
34915 |
529.1 |
59.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.415 ± 0.016 | 2.349 ± 0.016 |
4.888 ± 0.011 | 7.025 ± 0.022 |
3.783 ± 0.011 | 6.127 ± 0.026 |
2.607 ± 0.01 | 4.925 ± 0.012 |
6.281 ± 0.02 | 9.67 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.256 ± 0.008 | 4.077 ± 0.012 |
5.537 ± 0.026 | 4.683 ± 0.017 |
5.354 ± 0.016 | 8.269 ± 0.023 |
5.464 ± 0.017 | 6.122 ± 0.018 |
1.24 ± 0.006 | 2.861 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
