
Trametes pubescens (White-rot fungus)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Polyporales; Polyporaceae; Trametes
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
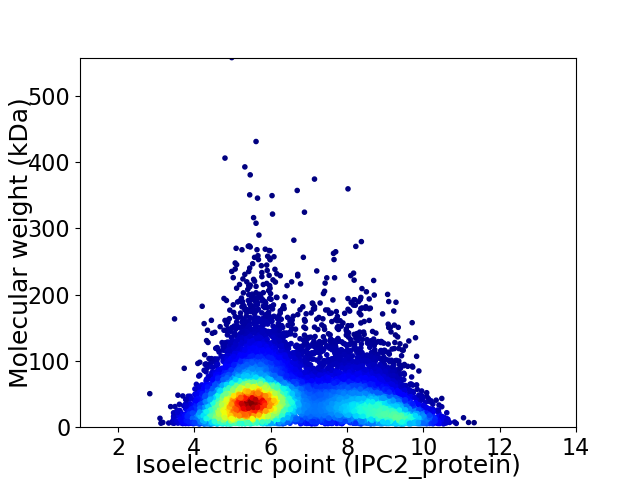
Virtual 2D-PAGE plot for 14147 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M2V285|A0A1M2V285_TRAPU Zinc finger protein 2 OS=Trametes pubescens OX=154538 GN=TRAPUB_7742 PE=4 SV=1
MM1 pKa = 8.4SMIMAEE7 pKa = 4.25SFDD10 pKa = 4.95APMTDD15 pKa = 4.23SGDD18 pKa = 3.82TDD20 pKa = 3.26VSMYY24 pKa = 9.66PGGMTADD31 pKa = 3.31SWLSAEE37 pKa = 4.38ASMGDD42 pKa = 3.45VAYY45 pKa = 10.74SADD48 pKa = 3.74TATFDD53 pKa = 3.56YY54 pKa = 9.29MQEE57 pKa = 4.0GVEE60 pKa = 4.8VEE62 pKa = 4.31MLDD65 pKa = 5.78DD66 pKa = 5.42LDD68 pKa = 6.08DD69 pKa = 4.72DD70 pKa = 4.51AAITEE75 pKa = 4.43YY76 pKa = 11.32EE77 pKa = 4.19MADD80 pKa = 4.28DD81 pKa = 4.28GDD83 pKa = 3.88GYY85 pKa = 11.19YY86 pKa = 10.71DD87 pKa = 4.36HH88 pKa = 7.3EE89 pKa = 5.77LEE91 pKa = 4.42DD92 pKa = 4.24VEE94 pKa = 4.57VLDD97 pKa = 4.02VSRR100 pKa = 11.84GASIPPVGPADD111 pKa = 3.49VSVDD115 pKa = 3.39ANAGNSEE122 pKa = 4.14MMHH125 pKa = 5.64VPPYY129 pKa = 10.82DD130 pKa = 3.52GGAPVASGSHH140 pKa = 5.33YY141 pKa = 10.74SVEE144 pKa = 4.01PSRR147 pKa = 11.84DD148 pKa = 3.27DD149 pKa = 3.27SALYY153 pKa = 10.8SEE155 pKa = 4.79THH157 pKa = 5.12STAHH161 pKa = 6.95PEE163 pKa = 4.23HH164 pKa = 6.85PVEE167 pKa = 5.31ADD169 pKa = 3.13ALAEE173 pKa = 4.03TSAPLEE179 pKa = 4.36GDD181 pKa = 3.35VHH183 pKa = 6.61HH184 pKa = 7.01AVSASSPSAPDD195 pKa = 3.01VSAVVEE201 pKa = 4.54DD202 pKa = 3.91NGAPAEE208 pKa = 4.24LPAFPTLNHH217 pKa = 6.88AEE219 pKa = 4.22DD220 pKa = 4.71ASTSTDD226 pKa = 3.29PAASEE231 pKa = 4.18SHH233 pKa = 6.27SVSEE237 pKa = 5.5DD238 pKa = 3.24RR239 pKa = 11.84LLDD242 pKa = 3.75PVLAGPSDD250 pKa = 3.87VDD252 pKa = 3.36HH253 pKa = 7.1TSPVVPVDD261 pKa = 3.95SADD264 pKa = 3.52LHH266 pKa = 6.36PGEE269 pKa = 4.78EE270 pKa = 4.56SAHH273 pKa = 5.96EE274 pKa = 3.94LAADD278 pKa = 4.19DD279 pKa = 3.99PHH281 pKa = 6.85EE282 pKa = 4.69ISDD285 pKa = 4.13GVYY288 pKa = 9.65IDD290 pKa = 4.54PPPPVLLSLPPSAEE304 pKa = 3.75HH305 pKa = 7.53AEE307 pKa = 4.15YY308 pKa = 11.07CLFNQPQSSTSPSTPGGSKK327 pKa = 10.06SEE329 pKa = 4.2PAAEE333 pKa = 4.04PLRR336 pKa = 11.84LLLHH340 pKa = 6.8DD341 pKa = 5.16RR342 pKa = 11.84PTLYY346 pKa = 10.87YY347 pKa = 9.97EE348 pKa = 4.35PLSSVFDD355 pKa = 3.96ALRR358 pKa = 11.84HH359 pKa = 5.18EE360 pKa = 4.41EE361 pKa = 4.81CILNLPGSSDD371 pKa = 3.61AEE373 pKa = 4.27LVLDD377 pKa = 4.72AYY379 pKa = 8.98EE380 pKa = 4.49LRR382 pKa = 11.84LAICEE387 pKa = 4.26DD388 pKa = 3.15NVYY391 pKa = 10.55AHH393 pKa = 6.92EE394 pKa = 4.45VTLHH398 pKa = 5.75EE399 pKa = 5.24LDD401 pKa = 4.53VIHH404 pKa = 6.95GGSDD408 pKa = 2.92LHH410 pKa = 7.38GPLRR414 pKa = 11.84LKK416 pKa = 10.86LKK418 pKa = 10.03FVAPRR423 pKa = 11.84FVTRR427 pKa = 11.84YY428 pKa = 9.24HH429 pKa = 7.38LLRR432 pKa = 11.84EE433 pKa = 4.23QISRR437 pKa = 11.84LNLTADD443 pKa = 3.51GDD445 pKa = 4.14EE446 pKa = 4.54LLSGVAASDD455 pKa = 3.34PVYY458 pKa = 9.9EE459 pKa = 3.95QEE461 pKa = 3.99YY462 pKa = 10.26DD463 pKa = 3.47VSHH466 pKa = 6.19EE467 pKa = 4.07AEE469 pKa = 4.64VPHH472 pKa = 6.26HH473 pKa = 6.11HH474 pKa = 7.18AEE476 pKa = 4.35DD477 pKa = 4.34GEE479 pKa = 4.2ARR481 pKa = 11.84ASVDD485 pKa = 3.09EE486 pKa = 5.47HH487 pKa = 7.7YY488 pKa = 10.9DD489 pKa = 3.56GPIEE493 pKa = 4.1GTEE496 pKa = 4.2DD497 pKa = 3.39GTHH500 pKa = 5.89EE501 pKa = 4.18EE502 pKa = 4.09LRR504 pKa = 11.84EE505 pKa = 4.08GVDD508 pKa = 3.21ASEE511 pKa = 4.99AEE513 pKa = 4.14ALAPNAGLTDD523 pKa = 3.21NHH525 pKa = 6.2VASEE529 pKa = 4.01RR530 pKa = 11.84HH531 pKa = 4.92EE532 pKa = 4.32AVPEE536 pKa = 3.93QLDD539 pKa = 4.24GEE541 pKa = 4.94DD542 pKa = 3.17GTAALEE548 pKa = 4.76GPSRR552 pKa = 11.84EE553 pKa = 4.4ATTAPPDD560 pKa = 3.37VFEE563 pKa = 5.3AEE565 pKa = 4.3RR566 pKa = 11.84ADD568 pKa = 3.92LEE570 pKa = 4.6HH571 pKa = 6.69EE572 pKa = 4.43HH573 pKa = 5.94EE574 pKa = 4.28QEE576 pKa = 4.53DD577 pKa = 3.92VGEE580 pKa = 4.68GEE582 pKa = 4.47GTLDD586 pKa = 3.49ANDD589 pKa = 3.57VHH591 pKa = 6.21EE592 pKa = 4.99HH593 pKa = 6.42ADD595 pKa = 3.3AGEE598 pKa = 4.31GGDD601 pKa = 3.75YY602 pKa = 10.92VEE604 pKa = 5.7HH605 pKa = 7.93DD606 pKa = 4.59EE607 pKa = 5.88FPDD610 pKa = 5.07DD611 pKa = 4.64EE612 pKa = 6.92DD613 pKa = 6.23DD614 pKa = 5.63FGDD617 pKa = 4.96DD618 pKa = 3.72LPEE621 pKa = 4.69EE622 pKa = 4.32IGGQTEE628 pKa = 4.13NEE630 pKa = 4.57AYY632 pKa = 8.05THH634 pKa = 6.35TGAVEE639 pKa = 4.17DD640 pKa = 4.69APQDD644 pKa = 3.27SAYY647 pKa = 10.6AEE649 pKa = 4.23EE650 pKa = 4.69EE651 pKa = 4.27GVDD654 pKa = 3.43AHH656 pKa = 7.09EE657 pKa = 5.31DD658 pKa = 3.41ALEE661 pKa = 4.25ASSGDD666 pKa = 3.72AGQGDD671 pKa = 4.54PDD673 pKa = 4.15AASLAQDD680 pKa = 4.32DD681 pKa = 4.9EE682 pKa = 5.72GPLADD687 pKa = 4.95GLDD690 pKa = 4.34TNGHH694 pKa = 5.96TDD696 pKa = 3.27TFAHH700 pKa = 7.03LDD702 pKa = 3.51SADD705 pKa = 4.57DD706 pKa = 3.75YY707 pKa = 12.03DD708 pKa = 4.27EE709 pKa = 5.94LHH711 pKa = 6.5LQDD714 pKa = 4.64VGTGQHH720 pKa = 6.87DD721 pKa = 4.38DD722 pKa = 3.7EE723 pKa = 4.88STAAHH728 pKa = 6.73PVTNEE733 pKa = 3.15HH734 pKa = 5.58SQNYY738 pKa = 9.43EE739 pKa = 3.84DD740 pKa = 5.92DD741 pKa = 4.16PLTNEE746 pKa = 4.0EE747 pKa = 4.47STTLSSEE754 pKa = 4.04EE755 pKa = 5.11DD756 pKa = 3.56GLADD760 pKa = 5.69DD761 pKa = 5.21WDD763 pKa = 5.26DD764 pKa = 5.28DD765 pKa = 5.58DD766 pKa = 6.82DD767 pKa = 4.65VLDD770 pKa = 3.98VDD772 pKa = 5.13AVPAPEE778 pKa = 4.44VNQADD783 pKa = 3.77ALSRR787 pKa = 11.84KK788 pKa = 9.57SSSATLASKK797 pKa = 10.4SSKK800 pKa = 8.78RR801 pKa = 11.84TYY803 pKa = 11.19DD804 pKa = 3.38EE805 pKa = 4.94VEE807 pKa = 4.54LDD809 pKa = 5.51DD810 pKa = 6.38FDD812 pKa = 6.89DD813 pKa = 6.13DD814 pKa = 4.55PSQLEE819 pKa = 4.19AASTPAEE826 pKa = 4.55DD827 pKa = 4.37VPSGPPQQ834 pKa = 3.39
MM1 pKa = 8.4SMIMAEE7 pKa = 4.25SFDD10 pKa = 4.95APMTDD15 pKa = 4.23SGDD18 pKa = 3.82TDD20 pKa = 3.26VSMYY24 pKa = 9.66PGGMTADD31 pKa = 3.31SWLSAEE37 pKa = 4.38ASMGDD42 pKa = 3.45VAYY45 pKa = 10.74SADD48 pKa = 3.74TATFDD53 pKa = 3.56YY54 pKa = 9.29MQEE57 pKa = 4.0GVEE60 pKa = 4.8VEE62 pKa = 4.31MLDD65 pKa = 5.78DD66 pKa = 5.42LDD68 pKa = 6.08DD69 pKa = 4.72DD70 pKa = 4.51AAITEE75 pKa = 4.43YY76 pKa = 11.32EE77 pKa = 4.19MADD80 pKa = 4.28DD81 pKa = 4.28GDD83 pKa = 3.88GYY85 pKa = 11.19YY86 pKa = 10.71DD87 pKa = 4.36HH88 pKa = 7.3EE89 pKa = 5.77LEE91 pKa = 4.42DD92 pKa = 4.24VEE94 pKa = 4.57VLDD97 pKa = 4.02VSRR100 pKa = 11.84GASIPPVGPADD111 pKa = 3.49VSVDD115 pKa = 3.39ANAGNSEE122 pKa = 4.14MMHH125 pKa = 5.64VPPYY129 pKa = 10.82DD130 pKa = 3.52GGAPVASGSHH140 pKa = 5.33YY141 pKa = 10.74SVEE144 pKa = 4.01PSRR147 pKa = 11.84DD148 pKa = 3.27DD149 pKa = 3.27SALYY153 pKa = 10.8SEE155 pKa = 4.79THH157 pKa = 5.12STAHH161 pKa = 6.95PEE163 pKa = 4.23HH164 pKa = 6.85PVEE167 pKa = 5.31ADD169 pKa = 3.13ALAEE173 pKa = 4.03TSAPLEE179 pKa = 4.36GDD181 pKa = 3.35VHH183 pKa = 6.61HH184 pKa = 7.01AVSASSPSAPDD195 pKa = 3.01VSAVVEE201 pKa = 4.54DD202 pKa = 3.91NGAPAEE208 pKa = 4.24LPAFPTLNHH217 pKa = 6.88AEE219 pKa = 4.22DD220 pKa = 4.71ASTSTDD226 pKa = 3.29PAASEE231 pKa = 4.18SHH233 pKa = 6.27SVSEE237 pKa = 5.5DD238 pKa = 3.24RR239 pKa = 11.84LLDD242 pKa = 3.75PVLAGPSDD250 pKa = 3.87VDD252 pKa = 3.36HH253 pKa = 7.1TSPVVPVDD261 pKa = 3.95SADD264 pKa = 3.52LHH266 pKa = 6.36PGEE269 pKa = 4.78EE270 pKa = 4.56SAHH273 pKa = 5.96EE274 pKa = 3.94LAADD278 pKa = 4.19DD279 pKa = 3.99PHH281 pKa = 6.85EE282 pKa = 4.69ISDD285 pKa = 4.13GVYY288 pKa = 9.65IDD290 pKa = 4.54PPPPVLLSLPPSAEE304 pKa = 3.75HH305 pKa = 7.53AEE307 pKa = 4.15YY308 pKa = 11.07CLFNQPQSSTSPSTPGGSKK327 pKa = 10.06SEE329 pKa = 4.2PAAEE333 pKa = 4.04PLRR336 pKa = 11.84LLLHH340 pKa = 6.8DD341 pKa = 5.16RR342 pKa = 11.84PTLYY346 pKa = 10.87YY347 pKa = 9.97EE348 pKa = 4.35PLSSVFDD355 pKa = 3.96ALRR358 pKa = 11.84HH359 pKa = 5.18EE360 pKa = 4.41EE361 pKa = 4.81CILNLPGSSDD371 pKa = 3.61AEE373 pKa = 4.27LVLDD377 pKa = 4.72AYY379 pKa = 8.98EE380 pKa = 4.49LRR382 pKa = 11.84LAICEE387 pKa = 4.26DD388 pKa = 3.15NVYY391 pKa = 10.55AHH393 pKa = 6.92EE394 pKa = 4.45VTLHH398 pKa = 5.75EE399 pKa = 5.24LDD401 pKa = 4.53VIHH404 pKa = 6.95GGSDD408 pKa = 2.92LHH410 pKa = 7.38GPLRR414 pKa = 11.84LKK416 pKa = 10.86LKK418 pKa = 10.03FVAPRR423 pKa = 11.84FVTRR427 pKa = 11.84YY428 pKa = 9.24HH429 pKa = 7.38LLRR432 pKa = 11.84EE433 pKa = 4.23QISRR437 pKa = 11.84LNLTADD443 pKa = 3.51GDD445 pKa = 4.14EE446 pKa = 4.54LLSGVAASDD455 pKa = 3.34PVYY458 pKa = 9.9EE459 pKa = 3.95QEE461 pKa = 3.99YY462 pKa = 10.26DD463 pKa = 3.47VSHH466 pKa = 6.19EE467 pKa = 4.07AEE469 pKa = 4.64VPHH472 pKa = 6.26HH473 pKa = 6.11HH474 pKa = 7.18AEE476 pKa = 4.35DD477 pKa = 4.34GEE479 pKa = 4.2ARR481 pKa = 11.84ASVDD485 pKa = 3.09EE486 pKa = 5.47HH487 pKa = 7.7YY488 pKa = 10.9DD489 pKa = 3.56GPIEE493 pKa = 4.1GTEE496 pKa = 4.2DD497 pKa = 3.39GTHH500 pKa = 5.89EE501 pKa = 4.18EE502 pKa = 4.09LRR504 pKa = 11.84EE505 pKa = 4.08GVDD508 pKa = 3.21ASEE511 pKa = 4.99AEE513 pKa = 4.14ALAPNAGLTDD523 pKa = 3.21NHH525 pKa = 6.2VASEE529 pKa = 4.01RR530 pKa = 11.84HH531 pKa = 4.92EE532 pKa = 4.32AVPEE536 pKa = 3.93QLDD539 pKa = 4.24GEE541 pKa = 4.94DD542 pKa = 3.17GTAALEE548 pKa = 4.76GPSRR552 pKa = 11.84EE553 pKa = 4.4ATTAPPDD560 pKa = 3.37VFEE563 pKa = 5.3AEE565 pKa = 4.3RR566 pKa = 11.84ADD568 pKa = 3.92LEE570 pKa = 4.6HH571 pKa = 6.69EE572 pKa = 4.43HH573 pKa = 5.94EE574 pKa = 4.28QEE576 pKa = 4.53DD577 pKa = 3.92VGEE580 pKa = 4.68GEE582 pKa = 4.47GTLDD586 pKa = 3.49ANDD589 pKa = 3.57VHH591 pKa = 6.21EE592 pKa = 4.99HH593 pKa = 6.42ADD595 pKa = 3.3AGEE598 pKa = 4.31GGDD601 pKa = 3.75YY602 pKa = 10.92VEE604 pKa = 5.7HH605 pKa = 7.93DD606 pKa = 4.59EE607 pKa = 5.88FPDD610 pKa = 5.07DD611 pKa = 4.64EE612 pKa = 6.92DD613 pKa = 6.23DD614 pKa = 5.63FGDD617 pKa = 4.96DD618 pKa = 3.72LPEE621 pKa = 4.69EE622 pKa = 4.32IGGQTEE628 pKa = 4.13NEE630 pKa = 4.57AYY632 pKa = 8.05THH634 pKa = 6.35TGAVEE639 pKa = 4.17DD640 pKa = 4.69APQDD644 pKa = 3.27SAYY647 pKa = 10.6AEE649 pKa = 4.23EE650 pKa = 4.69EE651 pKa = 4.27GVDD654 pKa = 3.43AHH656 pKa = 7.09EE657 pKa = 5.31DD658 pKa = 3.41ALEE661 pKa = 4.25ASSGDD666 pKa = 3.72AGQGDD671 pKa = 4.54PDD673 pKa = 4.15AASLAQDD680 pKa = 4.32DD681 pKa = 4.9EE682 pKa = 5.72GPLADD687 pKa = 4.95GLDD690 pKa = 4.34TNGHH694 pKa = 5.96TDD696 pKa = 3.27TFAHH700 pKa = 7.03LDD702 pKa = 3.51SADD705 pKa = 4.57DD706 pKa = 3.75YY707 pKa = 12.03DD708 pKa = 4.27EE709 pKa = 5.94LHH711 pKa = 6.5LQDD714 pKa = 4.64VGTGQHH720 pKa = 6.87DD721 pKa = 4.38DD722 pKa = 3.7EE723 pKa = 4.88STAAHH728 pKa = 6.73PVTNEE733 pKa = 3.15HH734 pKa = 5.58SQNYY738 pKa = 9.43EE739 pKa = 3.84DD740 pKa = 5.92DD741 pKa = 4.16PLTNEE746 pKa = 4.0EE747 pKa = 4.47STTLSSEE754 pKa = 4.04EE755 pKa = 5.11DD756 pKa = 3.56GLADD760 pKa = 5.69DD761 pKa = 5.21WDD763 pKa = 5.26DD764 pKa = 5.28DD765 pKa = 5.58DD766 pKa = 6.82DD767 pKa = 4.65VLDD770 pKa = 3.98VDD772 pKa = 5.13AVPAPEE778 pKa = 4.44VNQADD783 pKa = 3.77ALSRR787 pKa = 11.84KK788 pKa = 9.57SSSATLASKK797 pKa = 10.4SSKK800 pKa = 8.78RR801 pKa = 11.84TYY803 pKa = 11.19DD804 pKa = 3.38EE805 pKa = 4.94VEE807 pKa = 4.54LDD809 pKa = 5.51DD810 pKa = 6.38FDD812 pKa = 6.89DD813 pKa = 6.13DD814 pKa = 4.55PSQLEE819 pKa = 4.19AASTPAEE826 pKa = 4.55DD827 pKa = 4.37VPSGPPQQ834 pKa = 3.39
Molecular weight: 88.77 kDa
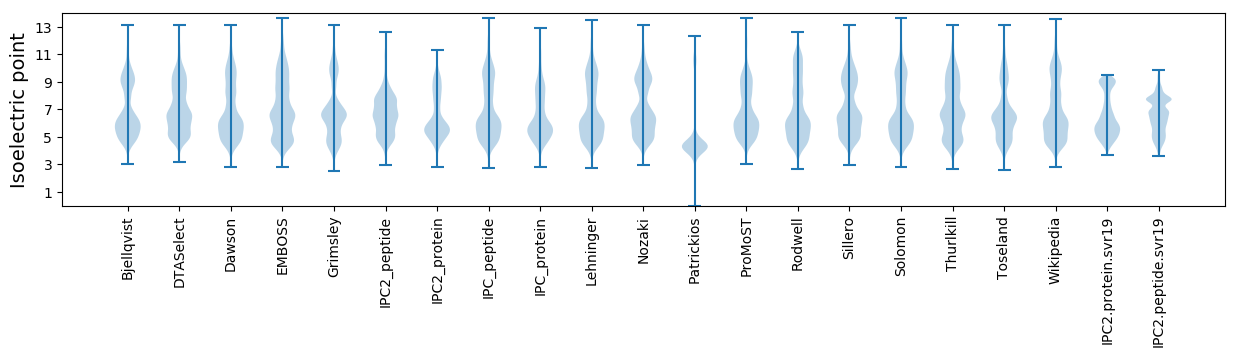
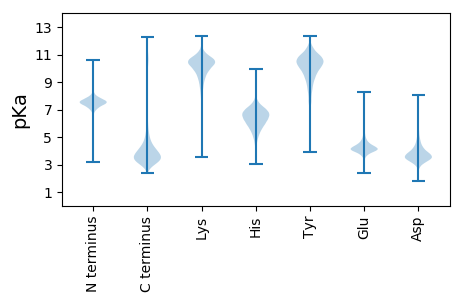
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M2W7U4|A0A1M2W7U4_TRAPU Uncharacterized protein OS=Trametes pubescens OX=154538 GN=TRAPUB_14140 PE=4 SV=1
AA1 pKa = 7.18TPAHH5 pKa = 6.54ASLRR9 pKa = 11.84PPNPRR14 pKa = 11.84LRR16 pKa = 11.84NRR18 pKa = 11.84ATQPRR23 pKa = 11.84PPAPSVPRR31 pKa = 11.84PAPRR35 pKa = 11.84APHH38 pKa = 5.24LHH40 pKa = 5.78RR41 pKa = 11.84RR42 pKa = 11.84ARR44 pKa = 11.84PHH46 pKa = 6.09VSHH49 pKa = 6.94SNRR52 pKa = 11.84AIRR55 pKa = 11.84LRR57 pKa = 11.84RR58 pKa = 11.84AAPP61 pKa = 3.36
AA1 pKa = 7.18TPAHH5 pKa = 6.54ASLRR9 pKa = 11.84PPNPRR14 pKa = 11.84LRR16 pKa = 11.84NRR18 pKa = 11.84ATQPRR23 pKa = 11.84PPAPSVPRR31 pKa = 11.84PAPRR35 pKa = 11.84APHH38 pKa = 5.24LHH40 pKa = 5.78RR41 pKa = 11.84RR42 pKa = 11.84ARR44 pKa = 11.84PHH46 pKa = 6.09VSHH49 pKa = 6.94SNRR52 pKa = 11.84AIRR55 pKa = 11.84LRR57 pKa = 11.84RR58 pKa = 11.84AAPP61 pKa = 3.36
Molecular weight: 6.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6007546 |
49 |
5047 |
424.7 |
46.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.985 ± 0.023 | 1.19 ± 0.009 |
5.779 ± 0.015 | 5.995 ± 0.02 |
3.474 ± 0.012 | 6.648 ± 0.021 |
2.6 ± 0.01 | 4.182 ± 0.013 |
4.106 ± 0.019 | 9.071 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.037 ± 0.008 | 2.94 ± 0.01 |
7.06 ± 0.029 | 3.645 ± 0.011 |
6.673 ± 0.018 | 8.144 ± 0.03 |
5.95 ± 0.014 | 6.522 ± 0.017 |
1.451 ± 0.008 | 2.546 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
