
Labilibaculum antarcticum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinifilaceae; Labilibaculum
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
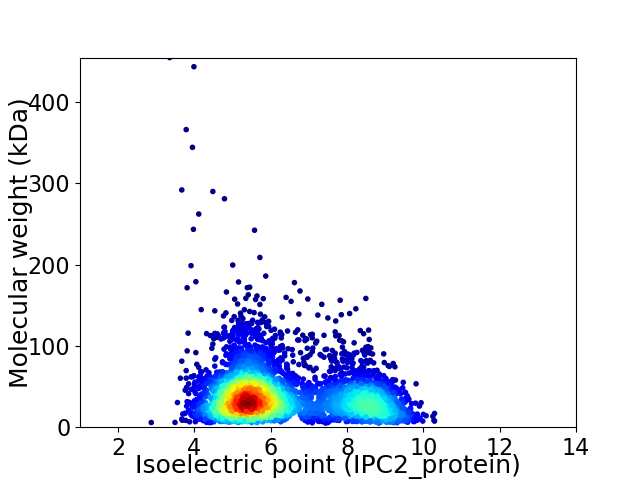
Virtual 2D-PAGE plot for 4329 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y1CMJ4|A0A1Y1CMJ4_9BACT Uncharacterized protein OS=Labilibaculum antarcticum OX=1717717 GN=ALGA_2938 PE=4 SV=1
MM1 pKa = 7.63RR2 pKa = 11.84LNWILISSLALLASTFACSDD22 pKa = 3.9DD23 pKa = 3.77DD24 pKa = 4.53TISDD28 pKa = 4.17SEE30 pKa = 4.68NITEE34 pKa = 4.21NLPDD38 pKa = 3.34ISGYY42 pKa = 10.0PIVGTNQSTAFNNTTNISTPSSGDD66 pKa = 3.17DD67 pKa = 3.97FYY69 pKa = 11.81GQNANYY75 pKa = 9.74PGTTPQYY82 pKa = 10.96VDD84 pKa = 3.81NGDD87 pKa = 3.62GTVTDD92 pKa = 4.2MVTGLMWQNTLDD104 pKa = 3.93HH105 pKa = 6.84NSDD108 pKa = 3.27GLINYY113 pKa = 8.13DD114 pKa = 4.58DD115 pKa = 4.37KK116 pKa = 11.35LTYY119 pKa = 10.76AEE121 pKa = 4.21ILALASEE128 pKa = 4.85VATGDD133 pKa = 3.55YY134 pKa = 10.51TDD136 pKa = 3.57WRR138 pKa = 11.84VPTIKK143 pKa = 10.21EE144 pKa = 4.07QYY146 pKa = 9.78SLMMFSGRR154 pKa = 11.84DD155 pKa = 3.12ISGYY159 pKa = 10.41EE160 pKa = 4.09GTSTEE165 pKa = 4.07GLTPFINTDD174 pKa = 3.41YY175 pKa = 11.35FGFNYY180 pKa = 10.46GDD182 pKa = 3.26TDD184 pKa = 3.37AGEE187 pKa = 4.13RR188 pKa = 11.84LIDD191 pKa = 3.8VQCASTNVSVGNTEE205 pKa = 3.44IEE207 pKa = 4.27MVFGVNFADD216 pKa = 5.03GRR218 pKa = 11.84IKK220 pKa = 10.9GYY222 pKa = 8.32GTTMMGQAKK231 pKa = 9.38VFNYY235 pKa = 10.74LMVRR239 pKa = 11.84GNTTYY244 pKa = 11.33GEE246 pKa = 4.4NNFSDD251 pKa = 3.99NADD254 pKa = 3.45GTISDD259 pKa = 4.9LATDD263 pKa = 4.48LMWMQDD269 pKa = 3.68DD270 pKa = 4.4SDD272 pKa = 5.54EE273 pKa = 4.44GMNWQNALAYY283 pKa = 10.19AEE285 pKa = 4.29SFEE288 pKa = 4.37FAGRR292 pKa = 11.84SDD294 pKa = 3.04WRR296 pKa = 11.84LPSAKK301 pKa = 9.33EE302 pKa = 3.63LQSIVDD308 pKa = 3.83YY309 pKa = 11.07SRR311 pKa = 11.84SPQSSASAAINPIFNCTQITNEE333 pKa = 4.23AGEE336 pKa = 4.2VDD338 pKa = 4.32YY339 pKa = 10.71PFYY342 pKa = 10.69WSGTTHH348 pKa = 6.68ATTGNDD354 pKa = 3.35NNGGWGAYY362 pKa = 7.41VAFGRR367 pKa = 11.84AMGNEE372 pKa = 4.13STILPDD378 pKa = 3.95DD379 pKa = 3.81QNPPTEE385 pKa = 4.79GEE387 pKa = 4.16QGPPPTDD394 pKa = 4.26NIDD397 pKa = 3.56SSSDD401 pKa = 3.61SINWTDD407 pKa = 3.16VHH409 pKa = 6.46GAGAQRR415 pKa = 11.84SDD417 pKa = 4.21PKK419 pKa = 11.08SGDD422 pKa = 3.22PSEE425 pKa = 4.3YY426 pKa = 10.64SEE428 pKa = 4.22GHH430 pKa = 6.1GPQGDD435 pKa = 3.42AVRR438 pKa = 11.84ILNYY442 pKa = 9.57IRR444 pKa = 11.84LVRR447 pKa = 11.84DD448 pKa = 4.02TKK450 pKa = 11.28
MM1 pKa = 7.63RR2 pKa = 11.84LNWILISSLALLASTFACSDD22 pKa = 3.9DD23 pKa = 3.77DD24 pKa = 4.53TISDD28 pKa = 4.17SEE30 pKa = 4.68NITEE34 pKa = 4.21NLPDD38 pKa = 3.34ISGYY42 pKa = 10.0PIVGTNQSTAFNNTTNISTPSSGDD66 pKa = 3.17DD67 pKa = 3.97FYY69 pKa = 11.81GQNANYY75 pKa = 9.74PGTTPQYY82 pKa = 10.96VDD84 pKa = 3.81NGDD87 pKa = 3.62GTVTDD92 pKa = 4.2MVTGLMWQNTLDD104 pKa = 3.93HH105 pKa = 6.84NSDD108 pKa = 3.27GLINYY113 pKa = 8.13DD114 pKa = 4.58DD115 pKa = 4.37KK116 pKa = 11.35LTYY119 pKa = 10.76AEE121 pKa = 4.21ILALASEE128 pKa = 4.85VATGDD133 pKa = 3.55YY134 pKa = 10.51TDD136 pKa = 3.57WRR138 pKa = 11.84VPTIKK143 pKa = 10.21EE144 pKa = 4.07QYY146 pKa = 9.78SLMMFSGRR154 pKa = 11.84DD155 pKa = 3.12ISGYY159 pKa = 10.41EE160 pKa = 4.09GTSTEE165 pKa = 4.07GLTPFINTDD174 pKa = 3.41YY175 pKa = 11.35FGFNYY180 pKa = 10.46GDD182 pKa = 3.26TDD184 pKa = 3.37AGEE187 pKa = 4.13RR188 pKa = 11.84LIDD191 pKa = 3.8VQCASTNVSVGNTEE205 pKa = 3.44IEE207 pKa = 4.27MVFGVNFADD216 pKa = 5.03GRR218 pKa = 11.84IKK220 pKa = 10.9GYY222 pKa = 8.32GTTMMGQAKK231 pKa = 9.38VFNYY235 pKa = 10.74LMVRR239 pKa = 11.84GNTTYY244 pKa = 11.33GEE246 pKa = 4.4NNFSDD251 pKa = 3.99NADD254 pKa = 3.45GTISDD259 pKa = 4.9LATDD263 pKa = 4.48LMWMQDD269 pKa = 3.68DD270 pKa = 4.4SDD272 pKa = 5.54EE273 pKa = 4.44GMNWQNALAYY283 pKa = 10.19AEE285 pKa = 4.29SFEE288 pKa = 4.37FAGRR292 pKa = 11.84SDD294 pKa = 3.04WRR296 pKa = 11.84LPSAKK301 pKa = 9.33EE302 pKa = 3.63LQSIVDD308 pKa = 3.83YY309 pKa = 11.07SRR311 pKa = 11.84SPQSSASAAINPIFNCTQITNEE333 pKa = 4.23AGEE336 pKa = 4.2VDD338 pKa = 4.32YY339 pKa = 10.71PFYY342 pKa = 10.69WSGTTHH348 pKa = 6.68ATTGNDD354 pKa = 3.35NNGGWGAYY362 pKa = 7.41VAFGRR367 pKa = 11.84AMGNEE372 pKa = 4.13STILPDD378 pKa = 3.95DD379 pKa = 3.81QNPPTEE385 pKa = 4.79GEE387 pKa = 4.16QGPPPTDD394 pKa = 4.26NIDD397 pKa = 3.56SSSDD401 pKa = 3.61SINWTDD407 pKa = 3.16VHH409 pKa = 6.46GAGAQRR415 pKa = 11.84SDD417 pKa = 4.21PKK419 pKa = 11.08SGDD422 pKa = 3.22PSEE425 pKa = 4.3YY426 pKa = 10.64SEE428 pKa = 4.22GHH430 pKa = 6.1GPQGDD435 pKa = 3.42AVRR438 pKa = 11.84ILNYY442 pKa = 9.57IRR444 pKa = 11.84LVRR447 pKa = 11.84DD448 pKa = 4.02TKK450 pKa = 11.28
Molecular weight: 48.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y1CGP5|A0A1Y1CGP5_9BACT 50S ribosomal protein L4 OS=Labilibaculum antarcticum OX=1717717 GN=rplD PE=3 SV=1
MM1 pKa = 7.23ARR3 pKa = 11.84IVGVDD8 pKa = 3.58LPQNKK13 pKa = 9.63RR14 pKa = 11.84GIISLTYY21 pKa = 8.96IYY23 pKa = 10.73GIGRR27 pKa = 11.84SAARR31 pKa = 11.84QILDD35 pKa = 3.62EE36 pKa = 5.04AEE38 pKa = 4.28VPHH41 pKa = 7.04DD42 pKa = 4.19KK43 pKa = 10.38QVKK46 pKa = 9.17DD47 pKa = 3.3WTDD50 pKa = 3.18EE51 pKa = 3.69EE52 pKa = 5.16SNRR55 pKa = 11.84IRR57 pKa = 11.84QAINANLKK65 pKa = 10.27VEE67 pKa = 4.2GEE69 pKa = 4.12LRR71 pKa = 11.84SLNQLNIKK79 pKa = 10.09RR80 pKa = 11.84LMDD83 pKa = 3.77IGCYY87 pKa = 9.44RR88 pKa = 11.84GIRR91 pKa = 11.84HH92 pKa = 7.01RR93 pKa = 11.84IGLPVRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 11.08NNSRR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 9.2GKK113 pKa = 10.5RR114 pKa = 11.84KK115 pKa = 7.64TVANKK120 pKa = 10.07KK121 pKa = 9.89KK122 pKa = 9.38ATKK125 pKa = 10.14
MM1 pKa = 7.23ARR3 pKa = 11.84IVGVDD8 pKa = 3.58LPQNKK13 pKa = 9.63RR14 pKa = 11.84GIISLTYY21 pKa = 8.96IYY23 pKa = 10.73GIGRR27 pKa = 11.84SAARR31 pKa = 11.84QILDD35 pKa = 3.62EE36 pKa = 5.04AEE38 pKa = 4.28VPHH41 pKa = 7.04DD42 pKa = 4.19KK43 pKa = 10.38QVKK46 pKa = 9.17DD47 pKa = 3.3WTDD50 pKa = 3.18EE51 pKa = 3.69EE52 pKa = 5.16SNRR55 pKa = 11.84IRR57 pKa = 11.84QAINANLKK65 pKa = 10.27VEE67 pKa = 4.2GEE69 pKa = 4.12LRR71 pKa = 11.84SLNQLNIKK79 pKa = 10.09RR80 pKa = 11.84LMDD83 pKa = 3.77IGCYY87 pKa = 9.44RR88 pKa = 11.84GIRR91 pKa = 11.84HH92 pKa = 7.01RR93 pKa = 11.84IGLPVRR99 pKa = 11.84GQRR102 pKa = 11.84TKK104 pKa = 11.08NNSRR108 pKa = 11.84TRR110 pKa = 11.84KK111 pKa = 9.2GKK113 pKa = 10.5RR114 pKa = 11.84KK115 pKa = 7.64TVANKK120 pKa = 10.07KK121 pKa = 9.89KK122 pKa = 9.38ATKK125 pKa = 10.14
Molecular weight: 14.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1607196 |
42 |
4359 |
371.3 |
41.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.234 ± 0.04 | 0.966 ± 0.013 |
5.691 ± 0.033 | 6.854 ± 0.038 |
5.067 ± 0.027 | 6.59 ± 0.041 |
1.757 ± 0.017 | 7.992 ± 0.036 |
7.706 ± 0.045 | 9.329 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.021 | 5.854 ± 0.035 |
3.295 ± 0.019 | 3.321 ± 0.02 |
3.555 ± 0.022 | 6.843 ± 0.037 |
5.173 ± 0.044 | 6.167 ± 0.031 |
1.153 ± 0.016 | 4.04 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
