
Capybara microvirus Cap3_SP_315
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
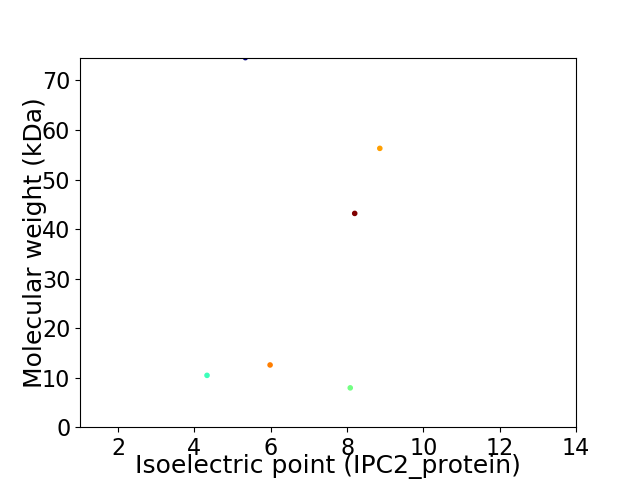
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W534|A0A4P8W534_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_315 OX=2585425 PE=4 SV=1
MM1 pKa = 7.41SFKK4 pKa = 10.89KK5 pKa = 10.75LIDD8 pKa = 3.7FQLPKK13 pKa = 10.43VDD15 pKa = 4.57DD16 pKa = 4.12PSILVHH22 pKa = 6.7EE23 pKa = 5.65DD24 pKa = 3.01IEE26 pKa = 4.59GTNFSRR32 pKa = 11.84LVEE35 pKa = 4.43KK36 pKa = 9.97PLSSIHH42 pKa = 6.91VDD44 pKa = 3.36LPDD47 pKa = 3.58FRR49 pKa = 11.84LVSLSEE55 pKa = 3.78ILSRR59 pKa = 11.84GEE61 pKa = 4.17DD62 pKa = 3.59LQKK65 pKa = 10.8VDD67 pKa = 3.51TKK69 pKa = 11.07LFEE72 pKa = 5.01SNLPDD77 pKa = 3.79FDD79 pKa = 4.93NDD81 pKa = 2.86IEE83 pKa = 4.67QIVNTYY89 pKa = 9.54EE90 pKa = 3.87SS91 pKa = 3.49
MM1 pKa = 7.41SFKK4 pKa = 10.89KK5 pKa = 10.75LIDD8 pKa = 3.7FQLPKK13 pKa = 10.43VDD15 pKa = 4.57DD16 pKa = 4.12PSILVHH22 pKa = 6.7EE23 pKa = 5.65DD24 pKa = 3.01IEE26 pKa = 4.59GTNFSRR32 pKa = 11.84LVEE35 pKa = 4.43KK36 pKa = 9.97PLSSIHH42 pKa = 6.91VDD44 pKa = 3.36LPDD47 pKa = 3.58FRR49 pKa = 11.84LVSLSEE55 pKa = 3.78ILSRR59 pKa = 11.84GEE61 pKa = 4.17DD62 pKa = 3.59LQKK65 pKa = 10.8VDD67 pKa = 3.51TKK69 pKa = 11.07LFEE72 pKa = 5.01SNLPDD77 pKa = 3.79FDD79 pKa = 4.93NDD81 pKa = 2.86IEE83 pKa = 4.67QIVNTYY89 pKa = 9.54EE90 pKa = 3.87SS91 pKa = 3.49
Molecular weight: 10.46 kDa
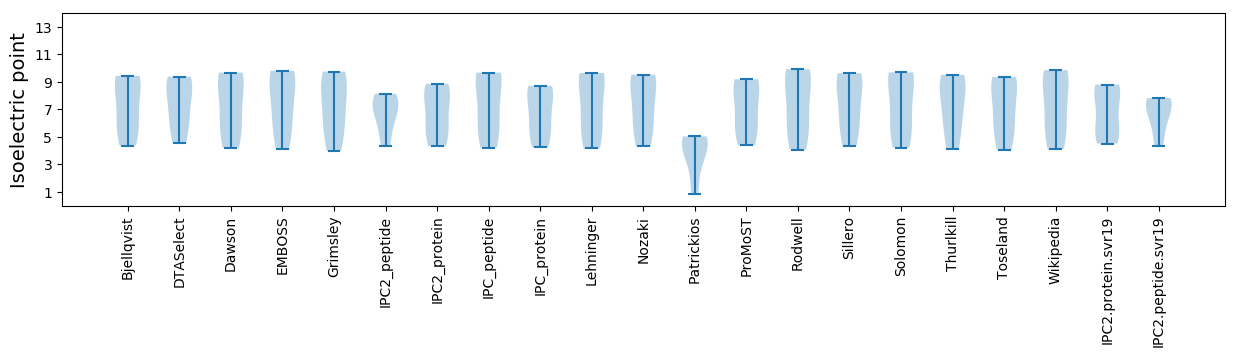
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8A8|A0A4P8W8A8_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_315 OX=2585425 PE=4 SV=1
MM1 pKa = 7.59CLSPIWIKK9 pKa = 11.12NPTRR13 pKa = 11.84DD14 pKa = 3.52YY15 pKa = 11.37RR16 pKa = 11.84PIDD19 pKa = 3.53KK20 pKa = 10.41VRR22 pKa = 11.84LQIPCGKK29 pKa = 10.03CPQCQQQKK37 pKa = 10.21AKK39 pKa = 10.47DD40 pKa = 3.57YY41 pKa = 8.92EE42 pKa = 4.11FRR44 pKa = 11.84MLSDD48 pKa = 3.97MYY50 pKa = 10.9RR51 pKa = 11.84KK52 pKa = 9.58GRR54 pKa = 11.84SFFFYY59 pKa = 10.34TLTYY63 pKa = 10.59NNQSLPYY70 pKa = 9.67RR71 pKa = 11.84EE72 pKa = 5.23FYY74 pKa = 10.53TLDD77 pKa = 3.69GEE79 pKa = 4.7IKK81 pKa = 10.29HH82 pKa = 5.77YY83 pKa = 9.33MCFVKK88 pKa = 10.61DD89 pKa = 3.83HH90 pKa = 6.95IKK92 pKa = 8.46THH94 pKa = 6.08INNIRR99 pKa = 11.84KK100 pKa = 8.96YY101 pKa = 8.24LHH103 pKa = 6.03KK104 pKa = 10.14HH105 pKa = 5.6FSFKK109 pKa = 10.66GCSFFCVPEE118 pKa = 3.96YY119 pKa = 11.36GDD121 pKa = 3.46LKK123 pKa = 11.07KK124 pKa = 10.27RR125 pKa = 11.84VHH127 pKa = 4.12WHH129 pKa = 6.01VYY131 pKa = 9.27FSTPVKK137 pKa = 10.3FDD139 pKa = 3.06AKK141 pKa = 10.59KK142 pKa = 10.2FLEE145 pKa = 4.11IVNRR149 pKa = 11.84FWNYY153 pKa = 9.19GLVFPEE159 pKa = 3.55HH160 pKa = 6.71HH161 pKa = 6.55YY162 pKa = 10.91SYY164 pKa = 9.84KK165 pKa = 6.95TTRR168 pKa = 11.84NRR170 pKa = 11.84LCRR173 pKa = 11.84GIEE176 pKa = 3.76IKK178 pKa = 10.93NPEE181 pKa = 3.89TSSYY185 pKa = 10.76YY186 pKa = 8.56IAHH189 pKa = 6.32YY190 pKa = 9.41VTDD193 pKa = 4.16FSIIRR198 pKa = 11.84DD199 pKa = 3.4IPHH202 pKa = 5.67VDD204 pKa = 2.86KK205 pKa = 11.33YY206 pKa = 11.81LNGLLLDD213 pKa = 4.73NDD215 pKa = 4.24TKK217 pKa = 11.29SYY219 pKa = 8.34TLRR222 pKa = 11.84PFVFMSKK229 pKa = 10.32SLGDD233 pKa = 3.64NEE235 pKa = 4.73EE236 pKa = 4.06LRR238 pKa = 11.84KK239 pKa = 10.71DD240 pKa = 3.4PDD242 pKa = 3.39KK243 pKa = 11.19LISYY247 pKa = 7.55FKK249 pKa = 10.52NGYY252 pKa = 6.82ITPFGKK258 pKa = 10.44KK259 pKa = 9.67LRR261 pKa = 11.84VPLFIIRR268 pKa = 11.84KK269 pKa = 5.85TLFDD273 pKa = 4.98IEE275 pKa = 4.44YY276 pKa = 9.66KK277 pKa = 11.02DD278 pKa = 4.43NGRR281 pKa = 11.84KK282 pKa = 9.7DD283 pKa = 3.75KK284 pKa = 11.26NNKK287 pKa = 8.91PSLDD291 pKa = 3.56VIYY294 pKa = 10.46KK295 pKa = 10.44LNSLGFKK302 pKa = 10.05FYY304 pKa = 11.08HH305 pKa = 6.08EE306 pKa = 4.67NFGNMIKK313 pKa = 10.3QKK315 pKa = 10.72AEE317 pKa = 3.77KK318 pKa = 9.21LAHH321 pKa = 6.82GFRR324 pKa = 11.84CMKK327 pKa = 10.32ALRR330 pKa = 11.84PQFFNIIEE338 pKa = 4.26KK339 pKa = 10.66KK340 pKa = 10.23FDD342 pKa = 3.37GSFEE346 pKa = 4.62DD347 pKa = 3.95KK348 pKa = 11.13LCINFFNDD356 pKa = 2.96KK357 pKa = 10.94QIFTKK362 pKa = 10.54FALYY366 pKa = 7.68DD367 pKa = 3.31TCFRR371 pKa = 11.84DD372 pKa = 3.75VKK374 pKa = 10.64SQNFMFGVPQSVKK387 pKa = 10.68DD388 pKa = 3.67LFRR391 pKa = 11.84ISPDD395 pKa = 2.5IYY397 pKa = 10.62LQRR400 pKa = 11.84KK401 pKa = 7.6LQYY404 pKa = 9.9KK405 pKa = 9.01PLIGKK410 pKa = 8.92SSPKK414 pKa = 9.94YY415 pKa = 9.67DD416 pKa = 3.5VKK418 pKa = 10.82EE419 pKa = 4.04TFSNYY424 pKa = 9.82RR425 pKa = 11.84EE426 pKa = 4.0FVFWDD431 pKa = 3.42YY432 pKa = 11.43CIDD435 pKa = 3.84SFNSCSARR443 pKa = 11.84TYY445 pKa = 10.58SRR447 pKa = 11.84SVFNLLEE454 pKa = 4.15FDD456 pKa = 4.34KK457 pKa = 10.97KK458 pKa = 11.17SQILDD463 pKa = 3.51LNYY466 pKa = 10.07VFF468 pKa = 5.92
MM1 pKa = 7.59CLSPIWIKK9 pKa = 11.12NPTRR13 pKa = 11.84DD14 pKa = 3.52YY15 pKa = 11.37RR16 pKa = 11.84PIDD19 pKa = 3.53KK20 pKa = 10.41VRR22 pKa = 11.84LQIPCGKK29 pKa = 10.03CPQCQQQKK37 pKa = 10.21AKK39 pKa = 10.47DD40 pKa = 3.57YY41 pKa = 8.92EE42 pKa = 4.11FRR44 pKa = 11.84MLSDD48 pKa = 3.97MYY50 pKa = 10.9RR51 pKa = 11.84KK52 pKa = 9.58GRR54 pKa = 11.84SFFFYY59 pKa = 10.34TLTYY63 pKa = 10.59NNQSLPYY70 pKa = 9.67RR71 pKa = 11.84EE72 pKa = 5.23FYY74 pKa = 10.53TLDD77 pKa = 3.69GEE79 pKa = 4.7IKK81 pKa = 10.29HH82 pKa = 5.77YY83 pKa = 9.33MCFVKK88 pKa = 10.61DD89 pKa = 3.83HH90 pKa = 6.95IKK92 pKa = 8.46THH94 pKa = 6.08INNIRR99 pKa = 11.84KK100 pKa = 8.96YY101 pKa = 8.24LHH103 pKa = 6.03KK104 pKa = 10.14HH105 pKa = 5.6FSFKK109 pKa = 10.66GCSFFCVPEE118 pKa = 3.96YY119 pKa = 11.36GDD121 pKa = 3.46LKK123 pKa = 11.07KK124 pKa = 10.27RR125 pKa = 11.84VHH127 pKa = 4.12WHH129 pKa = 6.01VYY131 pKa = 9.27FSTPVKK137 pKa = 10.3FDD139 pKa = 3.06AKK141 pKa = 10.59KK142 pKa = 10.2FLEE145 pKa = 4.11IVNRR149 pKa = 11.84FWNYY153 pKa = 9.19GLVFPEE159 pKa = 3.55HH160 pKa = 6.71HH161 pKa = 6.55YY162 pKa = 10.91SYY164 pKa = 9.84KK165 pKa = 6.95TTRR168 pKa = 11.84NRR170 pKa = 11.84LCRR173 pKa = 11.84GIEE176 pKa = 3.76IKK178 pKa = 10.93NPEE181 pKa = 3.89TSSYY185 pKa = 10.76YY186 pKa = 8.56IAHH189 pKa = 6.32YY190 pKa = 9.41VTDD193 pKa = 4.16FSIIRR198 pKa = 11.84DD199 pKa = 3.4IPHH202 pKa = 5.67VDD204 pKa = 2.86KK205 pKa = 11.33YY206 pKa = 11.81LNGLLLDD213 pKa = 4.73NDD215 pKa = 4.24TKK217 pKa = 11.29SYY219 pKa = 8.34TLRR222 pKa = 11.84PFVFMSKK229 pKa = 10.32SLGDD233 pKa = 3.64NEE235 pKa = 4.73EE236 pKa = 4.06LRR238 pKa = 11.84KK239 pKa = 10.71DD240 pKa = 3.4PDD242 pKa = 3.39KK243 pKa = 11.19LISYY247 pKa = 7.55FKK249 pKa = 10.52NGYY252 pKa = 6.82ITPFGKK258 pKa = 10.44KK259 pKa = 9.67LRR261 pKa = 11.84VPLFIIRR268 pKa = 11.84KK269 pKa = 5.85TLFDD273 pKa = 4.98IEE275 pKa = 4.44YY276 pKa = 9.66KK277 pKa = 11.02DD278 pKa = 4.43NGRR281 pKa = 11.84KK282 pKa = 9.7DD283 pKa = 3.75KK284 pKa = 11.26NNKK287 pKa = 8.91PSLDD291 pKa = 3.56VIYY294 pKa = 10.46KK295 pKa = 10.44LNSLGFKK302 pKa = 10.05FYY304 pKa = 11.08HH305 pKa = 6.08EE306 pKa = 4.67NFGNMIKK313 pKa = 10.3QKK315 pKa = 10.72AEE317 pKa = 3.77KK318 pKa = 9.21LAHH321 pKa = 6.82GFRR324 pKa = 11.84CMKK327 pKa = 10.32ALRR330 pKa = 11.84PQFFNIIEE338 pKa = 4.26KK339 pKa = 10.66KK340 pKa = 10.23FDD342 pKa = 3.37GSFEE346 pKa = 4.62DD347 pKa = 3.95KK348 pKa = 11.13LCINFFNDD356 pKa = 2.96KK357 pKa = 10.94QIFTKK362 pKa = 10.54FALYY366 pKa = 7.68DD367 pKa = 3.31TCFRR371 pKa = 11.84DD372 pKa = 3.75VKK374 pKa = 10.64SQNFMFGVPQSVKK387 pKa = 10.68DD388 pKa = 3.67LFRR391 pKa = 11.84ISPDD395 pKa = 2.5IYY397 pKa = 10.62LQRR400 pKa = 11.84KK401 pKa = 7.6LQYY404 pKa = 9.9KK405 pKa = 9.01PLIGKK410 pKa = 8.92SSPKK414 pKa = 9.94YY415 pKa = 9.67DD416 pKa = 3.5VKK418 pKa = 10.82EE419 pKa = 4.04TFSNYY424 pKa = 9.82RR425 pKa = 11.84EE426 pKa = 4.0FVFWDD431 pKa = 3.42YY432 pKa = 11.43CIDD435 pKa = 3.84SFNSCSARR443 pKa = 11.84TYY445 pKa = 10.58SRR447 pKa = 11.84SVFNLLEE454 pKa = 4.15FDD456 pKa = 4.34KK457 pKa = 10.97KK458 pKa = 11.17SQILDD463 pKa = 3.51LNYY466 pKa = 10.07VFF468 pKa = 5.92
Molecular weight: 56.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1784 |
68 |
659 |
297.3 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.372 ± 2.094 | 1.794 ± 0.463 |
6.67 ± 0.728 | 4.092 ± 0.528 |
6.502 ± 1.54 | 4.372 ± 0.723 |
2.074 ± 0.542 | 6.839 ± 0.546 |
7.287 ± 1.517 | 10.258 ± 1.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.289 ± 0.262 | 7.231 ± 0.959 |
3.98 ± 0.394 | 4.204 ± 0.701 |
4.092 ± 0.56 | 8.352 ± 0.65 |
6.11 ± 1.242 | 4.652 ± 0.327 |
0.841 ± 0.22 | 4.989 ± 0.954 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
