
Peanut chlorotic streak virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Soymovirus
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
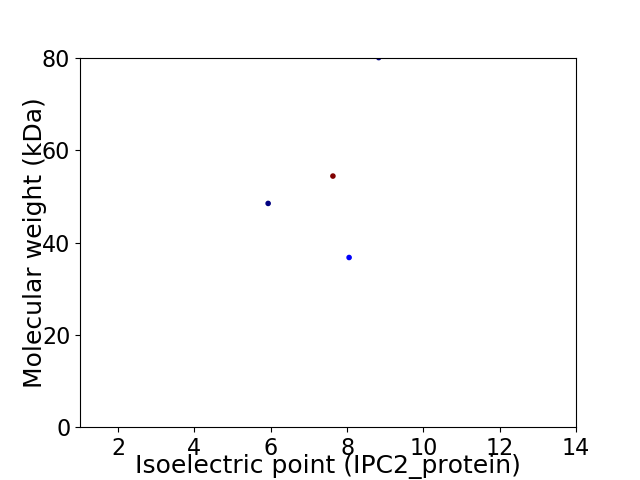
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q84684|Q84684_9VIRU Putative cell-to-cell transport protein OS=Peanut chlorotic streak virus OX=35593 PE=4 SV=1
MM1 pKa = 7.74EE2 pKa = 7.07DD3 pKa = 3.06IFKK6 pKa = 10.16MISEE10 pKa = 4.32KK11 pKa = 10.56KK12 pKa = 9.45EE13 pKa = 3.72KK14 pKa = 10.41LKK16 pKa = 11.18EE17 pKa = 4.05EE18 pKa = 4.11IEE20 pKa = 4.38EE21 pKa = 4.11KK22 pKa = 10.39RR23 pKa = 11.84SLVSRR28 pKa = 11.84LQKK31 pKa = 10.43EE32 pKa = 4.04ISDD35 pKa = 4.02KK36 pKa = 11.03EE37 pKa = 4.26KK38 pKa = 10.73EE39 pKa = 4.03LSALSQTEE47 pKa = 3.74QSLGLSIKK55 pKa = 10.4KK56 pKa = 7.79EE57 pKa = 3.99QPEE60 pKa = 4.04KK61 pKa = 11.0LVIQSSSEE69 pKa = 3.98KK70 pKa = 10.0YY71 pKa = 10.4DD72 pKa = 3.35EE73 pKa = 4.1EE74 pKa = 4.2EE75 pKa = 3.75RR76 pKa = 11.84AKK78 pKa = 10.24RR79 pKa = 11.84YY80 pKa = 9.16YY81 pKa = 10.63VIYY84 pKa = 9.98NGPGKK89 pKa = 10.54GIYY92 pKa = 9.82DD93 pKa = 3.61EE94 pKa = 4.26WGKK97 pKa = 11.08ASLFITGVKK106 pKa = 10.09GIRR109 pKa = 11.84HH110 pKa = 5.96KK111 pKa = 11.09KK112 pKa = 9.36FLSKK116 pKa = 10.83KK117 pKa = 8.13EE118 pKa = 4.02AQDD121 pKa = 3.9SFNEE125 pKa = 4.12EE126 pKa = 3.71NKK128 pKa = 10.12EE129 pKa = 3.86AAKK132 pKa = 9.11TAEE135 pKa = 3.86VSKK138 pKa = 10.93AYY140 pKa = 10.41LEE142 pKa = 4.46KK143 pKa = 10.71LQSPKK148 pKa = 10.22PQTSRR153 pKa = 11.84MVNLGKK159 pKa = 10.11IPSPRR164 pKa = 11.84TIDD167 pKa = 4.27HH168 pKa = 7.03ILTKK172 pKa = 10.6ADD174 pKa = 3.62LQEE177 pKa = 4.26KK178 pKa = 10.48ARR180 pKa = 11.84LTKK183 pKa = 10.31KK184 pKa = 10.34IYY186 pKa = 10.53DD187 pKa = 4.08DD188 pKa = 5.82DD189 pKa = 5.0YY190 pKa = 11.48MFLKK194 pKa = 10.36KK195 pKa = 10.9YY196 pKa = 10.82SDD198 pKa = 4.06DD199 pKa = 4.16DD200 pKa = 4.04KK201 pKa = 10.96QTSIYY206 pKa = 10.07PVDD209 pKa = 4.23SKK211 pKa = 11.88SLGAKK216 pKa = 9.82AVVLPEE222 pKa = 4.12ADD224 pKa = 3.58NIKK227 pKa = 10.02TYY229 pKa = 10.23MLYY232 pKa = 9.88QCGFIHH238 pKa = 8.03AIYY241 pKa = 9.91FKK243 pKa = 10.91SLDD246 pKa = 3.1IFRR249 pKa = 11.84YY250 pKa = 9.38FPEE253 pKa = 3.85NMKK256 pKa = 10.47RR257 pKa = 11.84AIQVYY262 pKa = 7.61AQRR265 pKa = 11.84MLKK268 pKa = 10.24AEE270 pKa = 3.8KK271 pKa = 10.48GMRR274 pKa = 11.84EE275 pKa = 3.96GFLRR279 pKa = 11.84IYY281 pKa = 9.06STNPEE286 pKa = 3.89FDD288 pKa = 3.08EE289 pKa = 4.69SGEE292 pKa = 4.25VVQPAIQLCQIGGSNSNYY310 pKa = 9.34PVMGVIEE317 pKa = 4.92DD318 pKa = 3.66VDD320 pKa = 3.58EE321 pKa = 5.35LEE323 pKa = 5.23AFLHH327 pKa = 6.11NYY329 pKa = 8.77TYY331 pKa = 10.97ILQKK335 pKa = 10.59GISVQGRR342 pKa = 11.84INYY345 pKa = 7.89SAKK348 pKa = 9.65STIIWSQQARR358 pKa = 11.84KK359 pKa = 9.14PSEE362 pKa = 3.55QDD364 pKa = 2.89IQILTEE370 pKa = 3.84FLLRR374 pKa = 11.84TVKK377 pKa = 10.95NEE379 pKa = 3.79FNFSAEE385 pKa = 4.25VKK387 pKa = 10.59QEE389 pKa = 3.18ICLRR393 pKa = 11.84LQRR396 pKa = 11.84DD397 pKa = 3.53FSEE400 pKa = 4.75DD401 pKa = 3.62HH402 pKa = 6.35VCQLCEE408 pKa = 3.87QSSGDD413 pKa = 3.62LEE415 pKa = 4.81PIKK418 pKa = 10.89EE419 pKa = 4.12EE420 pKa = 4.01
MM1 pKa = 7.74EE2 pKa = 7.07DD3 pKa = 3.06IFKK6 pKa = 10.16MISEE10 pKa = 4.32KK11 pKa = 10.56KK12 pKa = 9.45EE13 pKa = 3.72KK14 pKa = 10.41LKK16 pKa = 11.18EE17 pKa = 4.05EE18 pKa = 4.11IEE20 pKa = 4.38EE21 pKa = 4.11KK22 pKa = 10.39RR23 pKa = 11.84SLVSRR28 pKa = 11.84LQKK31 pKa = 10.43EE32 pKa = 4.04ISDD35 pKa = 4.02KK36 pKa = 11.03EE37 pKa = 4.26KK38 pKa = 10.73EE39 pKa = 4.03LSALSQTEE47 pKa = 3.74QSLGLSIKK55 pKa = 10.4KK56 pKa = 7.79EE57 pKa = 3.99QPEE60 pKa = 4.04KK61 pKa = 11.0LVIQSSSEE69 pKa = 3.98KK70 pKa = 10.0YY71 pKa = 10.4DD72 pKa = 3.35EE73 pKa = 4.1EE74 pKa = 4.2EE75 pKa = 3.75RR76 pKa = 11.84AKK78 pKa = 10.24RR79 pKa = 11.84YY80 pKa = 9.16YY81 pKa = 10.63VIYY84 pKa = 9.98NGPGKK89 pKa = 10.54GIYY92 pKa = 9.82DD93 pKa = 3.61EE94 pKa = 4.26WGKK97 pKa = 11.08ASLFITGVKK106 pKa = 10.09GIRR109 pKa = 11.84HH110 pKa = 5.96KK111 pKa = 11.09KK112 pKa = 9.36FLSKK116 pKa = 10.83KK117 pKa = 8.13EE118 pKa = 4.02AQDD121 pKa = 3.9SFNEE125 pKa = 4.12EE126 pKa = 3.71NKK128 pKa = 10.12EE129 pKa = 3.86AAKK132 pKa = 9.11TAEE135 pKa = 3.86VSKK138 pKa = 10.93AYY140 pKa = 10.41LEE142 pKa = 4.46KK143 pKa = 10.71LQSPKK148 pKa = 10.22PQTSRR153 pKa = 11.84MVNLGKK159 pKa = 10.11IPSPRR164 pKa = 11.84TIDD167 pKa = 4.27HH168 pKa = 7.03ILTKK172 pKa = 10.6ADD174 pKa = 3.62LQEE177 pKa = 4.26KK178 pKa = 10.48ARR180 pKa = 11.84LTKK183 pKa = 10.31KK184 pKa = 10.34IYY186 pKa = 10.53DD187 pKa = 4.08DD188 pKa = 5.82DD189 pKa = 5.0YY190 pKa = 11.48MFLKK194 pKa = 10.36KK195 pKa = 10.9YY196 pKa = 10.82SDD198 pKa = 4.06DD199 pKa = 4.16DD200 pKa = 4.04KK201 pKa = 10.96QTSIYY206 pKa = 10.07PVDD209 pKa = 4.23SKK211 pKa = 11.88SLGAKK216 pKa = 9.82AVVLPEE222 pKa = 4.12ADD224 pKa = 3.58NIKK227 pKa = 10.02TYY229 pKa = 10.23MLYY232 pKa = 9.88QCGFIHH238 pKa = 8.03AIYY241 pKa = 9.91FKK243 pKa = 10.91SLDD246 pKa = 3.1IFRR249 pKa = 11.84YY250 pKa = 9.38FPEE253 pKa = 3.85NMKK256 pKa = 10.47RR257 pKa = 11.84AIQVYY262 pKa = 7.61AQRR265 pKa = 11.84MLKK268 pKa = 10.24AEE270 pKa = 3.8KK271 pKa = 10.48GMRR274 pKa = 11.84EE275 pKa = 3.96GFLRR279 pKa = 11.84IYY281 pKa = 9.06STNPEE286 pKa = 3.89FDD288 pKa = 3.08EE289 pKa = 4.69SGEE292 pKa = 4.25VVQPAIQLCQIGGSNSNYY310 pKa = 9.34PVMGVIEE317 pKa = 4.92DD318 pKa = 3.66VDD320 pKa = 3.58EE321 pKa = 5.35LEE323 pKa = 5.23AFLHH327 pKa = 6.11NYY329 pKa = 8.77TYY331 pKa = 10.97ILQKK335 pKa = 10.59GISVQGRR342 pKa = 11.84INYY345 pKa = 7.89SAKK348 pKa = 9.65STIIWSQQARR358 pKa = 11.84KK359 pKa = 9.14PSEE362 pKa = 3.55QDD364 pKa = 2.89IQILTEE370 pKa = 3.84FLLRR374 pKa = 11.84TVKK377 pKa = 10.95NEE379 pKa = 3.79FNFSAEE385 pKa = 4.25VKK387 pKa = 10.59QEE389 pKa = 3.18ICLRR393 pKa = 11.84LQRR396 pKa = 11.84DD397 pKa = 3.53FSEE400 pKa = 4.75DD401 pKa = 3.62HH402 pKa = 6.35VCQLCEE408 pKa = 3.87QSSGDD413 pKa = 3.62LEE415 pKa = 4.81PIKK418 pKa = 10.89EE419 pKa = 4.12EE420 pKa = 4.01
Molecular weight: 48.52 kDa
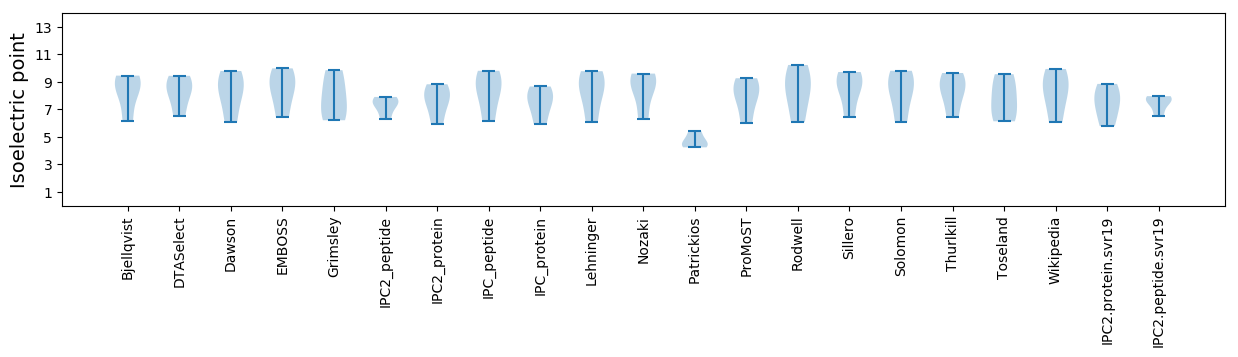
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q84683|Q84683_9VIRU Inclusion body matrix protein OS=Peanut chlorotic streak virus OX=35593 PE=3 SV=1
MM1 pKa = 7.4SSKK4 pKa = 10.39NSSFIKK10 pKa = 10.3VKK12 pKa = 10.72LFNKK16 pKa = 9.87YY17 pKa = 9.79LYY19 pKa = 10.6AYY21 pKa = 8.69IDD23 pKa = 3.55TGATICLAQAKK34 pKa = 9.36ILPIKK39 pKa = 9.34YY40 pKa = 7.09WKK42 pKa = 10.7KK43 pKa = 8.3MIKK46 pKa = 9.22PIKK49 pKa = 10.24VRR51 pKa = 11.84IANNKK56 pKa = 8.65VIHH59 pKa = 6.15IWYY62 pKa = 9.43KK63 pKa = 10.83AVDD66 pKa = 4.05LVLLLEE72 pKa = 4.96GKK74 pKa = 10.1RR75 pKa = 11.84FPLPSVYY82 pKa = 10.23QQDD85 pKa = 3.23AGLPLILGNNFLKK98 pKa = 10.69LYY100 pKa = 10.68NPFIQTLEE108 pKa = 4.37TISLRR113 pKa = 11.84CPQLEE118 pKa = 4.25KK119 pKa = 10.77QPSSLITTKK128 pKa = 10.17IYY130 pKa = 9.9NTFSLFGGVIVNILKK145 pKa = 8.76QQIYY149 pKa = 9.92IAIEE153 pKa = 4.13DD154 pKa = 4.16EE155 pKa = 4.4VTQLLEE161 pKa = 4.99AICSQNPLDD170 pKa = 3.89PQKK173 pKa = 11.03NRR175 pKa = 11.84NQIIVHH181 pKa = 6.7IDD183 pKa = 3.67LIDD186 pKa = 3.57PTKK189 pKa = 10.49EE190 pKa = 3.76VNVPNRR196 pKa = 11.84IPYY199 pKa = 5.78TQKK202 pKa = 10.96DD203 pKa = 2.86IDD205 pKa = 3.81EE206 pKa = 4.52FRR208 pKa = 11.84EE209 pKa = 4.15EE210 pKa = 3.76TSKK213 pKa = 11.08QIEE216 pKa = 4.26LGILRR221 pKa = 11.84QSKK224 pKa = 10.15SPHH227 pKa = 5.9SAPAFYY233 pKa = 10.93VEE235 pKa = 3.96NHH237 pKa = 5.76NEE239 pKa = 3.48IKK241 pKa = 10.17RR242 pKa = 11.84GKK244 pKa = 9.21RR245 pKa = 11.84RR246 pKa = 11.84LVINYY251 pKa = 9.15KK252 pKa = 8.7MNKK255 pKa = 7.06ATKK258 pKa = 9.82GDD260 pKa = 3.77AYY262 pKa = 11.35NLDD265 pKa = 3.65RR266 pKa = 11.84LYY268 pKa = 10.78LTDD271 pKa = 5.48RR272 pKa = 11.84EE273 pKa = 4.8SNWFSTLDD281 pKa = 3.42AKK283 pKa = 11.03SGFLQLRR290 pKa = 11.84LDD292 pKa = 4.26EE293 pKa = 4.27EE294 pKa = 4.85TKK296 pKa = 10.59PLTAFSCPPQMHH308 pKa = 6.83LEE310 pKa = 4.16YY311 pKa = 10.94NVMPMGLKK319 pKa = 9.34QAPSQFQRR327 pKa = 11.84FMDD330 pKa = 3.77NNLRR334 pKa = 11.84GLEE337 pKa = 4.71DD338 pKa = 3.16ISLAYY343 pKa = 9.58IDD345 pKa = 5.51DD346 pKa = 4.58IIVFTKK352 pKa = 9.22GTKK355 pKa = 9.64DD356 pKa = 3.32YY357 pKa = 10.9HH358 pKa = 6.46LKK360 pKa = 9.84QVARR364 pKa = 11.84VLIQLGNHH372 pKa = 5.35GVILSKK378 pKa = 10.84EE379 pKa = 3.81KK380 pKa = 10.98AKK382 pKa = 10.56IAFEE386 pKa = 4.34EE387 pKa = 4.55IEE389 pKa = 4.1FLGLKK394 pKa = 9.3ILKK397 pKa = 10.13NGFIEE402 pKa = 4.34PQKK405 pKa = 11.02HH406 pKa = 5.37LLEE409 pKa = 5.37KK410 pKa = 10.15IAEE413 pKa = 4.44FPDD416 pKa = 3.22QLQDD420 pKa = 2.92RR421 pKa = 11.84KK422 pKa = 10.55QIQKK426 pKa = 10.4FLGCLNYY433 pKa = 9.87IGEE436 pKa = 4.24KK437 pKa = 10.95GFFKK441 pKa = 10.68EE442 pKa = 3.76LAKK445 pKa = 10.27EE446 pKa = 4.02RR447 pKa = 11.84KK448 pKa = 9.07VLQKK452 pKa = 9.88MLSEE456 pKa = 4.03KK457 pKa = 10.8LPWKK461 pKa = 9.99WNDD464 pKa = 3.19LATLAVKK471 pKa = 10.36RR472 pKa = 11.84LKK474 pKa = 9.98QVCKK478 pKa = 10.14NLPRR482 pKa = 11.84LYY484 pKa = 10.09VAKK487 pKa = 10.36PSDD490 pKa = 4.22LLILTTDD497 pKa = 3.8ASDD500 pKa = 3.93TTWGAVLMAVPNAYY514 pKa = 9.9EE515 pKa = 4.01EE516 pKa = 4.31FLSFVTNNRR525 pKa = 11.84PSDD528 pKa = 3.38LKK530 pKa = 10.34TFLKK534 pKa = 10.46QKK536 pKa = 9.66EE537 pKa = 4.34GSSRR541 pKa = 11.84YY542 pKa = 9.41AEE544 pKa = 3.82KK545 pKa = 10.63SVPEE549 pKa = 4.6KK550 pKa = 10.64LNSMHH555 pKa = 7.21DD556 pKa = 3.36KK557 pKa = 10.82SHH559 pKa = 6.41GEE561 pKa = 3.77KK562 pKa = 10.49AEE564 pKa = 4.2KK565 pKa = 10.23SVSRR569 pKa = 11.84QTSQGRR575 pKa = 11.84SSSQSQQSYY584 pKa = 9.56IGSFMITKK592 pKa = 9.41WSSGTFKK599 pKa = 10.45PAEE602 pKa = 4.0EE603 pKa = 4.68NYY605 pKa = 9.09TVHH608 pKa = 6.7EE609 pKa = 4.49KK610 pKa = 10.86EE611 pKa = 4.29LLAFVNALKK620 pKa = 10.66KK621 pKa = 10.29FQIDD625 pKa = 3.5LRR627 pKa = 11.84PVKK630 pKa = 10.54FIFQTDD636 pKa = 3.66NSWVQGFLVNKK647 pKa = 10.34LKK649 pKa = 9.5EE650 pKa = 4.13TYY652 pKa = 10.3NRR654 pKa = 11.84GRR656 pKa = 11.84LARR659 pKa = 11.84WSMFIQQFDD668 pKa = 4.72FISLHH673 pKa = 5.62IAGKK677 pKa = 9.99EE678 pKa = 3.73NYY680 pKa = 9.95LADD683 pKa = 3.61TLTRR687 pKa = 11.84EE688 pKa = 4.4WKK690 pKa = 8.93TSSRR694 pKa = 3.78
MM1 pKa = 7.4SSKK4 pKa = 10.39NSSFIKK10 pKa = 10.3VKK12 pKa = 10.72LFNKK16 pKa = 9.87YY17 pKa = 9.79LYY19 pKa = 10.6AYY21 pKa = 8.69IDD23 pKa = 3.55TGATICLAQAKK34 pKa = 9.36ILPIKK39 pKa = 9.34YY40 pKa = 7.09WKK42 pKa = 10.7KK43 pKa = 8.3MIKK46 pKa = 9.22PIKK49 pKa = 10.24VRR51 pKa = 11.84IANNKK56 pKa = 8.65VIHH59 pKa = 6.15IWYY62 pKa = 9.43KK63 pKa = 10.83AVDD66 pKa = 4.05LVLLLEE72 pKa = 4.96GKK74 pKa = 10.1RR75 pKa = 11.84FPLPSVYY82 pKa = 10.23QQDD85 pKa = 3.23AGLPLILGNNFLKK98 pKa = 10.69LYY100 pKa = 10.68NPFIQTLEE108 pKa = 4.37TISLRR113 pKa = 11.84CPQLEE118 pKa = 4.25KK119 pKa = 10.77QPSSLITTKK128 pKa = 10.17IYY130 pKa = 9.9NTFSLFGGVIVNILKK145 pKa = 8.76QQIYY149 pKa = 9.92IAIEE153 pKa = 4.13DD154 pKa = 4.16EE155 pKa = 4.4VTQLLEE161 pKa = 4.99AICSQNPLDD170 pKa = 3.89PQKK173 pKa = 11.03NRR175 pKa = 11.84NQIIVHH181 pKa = 6.7IDD183 pKa = 3.67LIDD186 pKa = 3.57PTKK189 pKa = 10.49EE190 pKa = 3.76VNVPNRR196 pKa = 11.84IPYY199 pKa = 5.78TQKK202 pKa = 10.96DD203 pKa = 2.86IDD205 pKa = 3.81EE206 pKa = 4.52FRR208 pKa = 11.84EE209 pKa = 4.15EE210 pKa = 3.76TSKK213 pKa = 11.08QIEE216 pKa = 4.26LGILRR221 pKa = 11.84QSKK224 pKa = 10.15SPHH227 pKa = 5.9SAPAFYY233 pKa = 10.93VEE235 pKa = 3.96NHH237 pKa = 5.76NEE239 pKa = 3.48IKK241 pKa = 10.17RR242 pKa = 11.84GKK244 pKa = 9.21RR245 pKa = 11.84RR246 pKa = 11.84LVINYY251 pKa = 9.15KK252 pKa = 8.7MNKK255 pKa = 7.06ATKK258 pKa = 9.82GDD260 pKa = 3.77AYY262 pKa = 11.35NLDD265 pKa = 3.65RR266 pKa = 11.84LYY268 pKa = 10.78LTDD271 pKa = 5.48RR272 pKa = 11.84EE273 pKa = 4.8SNWFSTLDD281 pKa = 3.42AKK283 pKa = 11.03SGFLQLRR290 pKa = 11.84LDD292 pKa = 4.26EE293 pKa = 4.27EE294 pKa = 4.85TKK296 pKa = 10.59PLTAFSCPPQMHH308 pKa = 6.83LEE310 pKa = 4.16YY311 pKa = 10.94NVMPMGLKK319 pKa = 9.34QAPSQFQRR327 pKa = 11.84FMDD330 pKa = 3.77NNLRR334 pKa = 11.84GLEE337 pKa = 4.71DD338 pKa = 3.16ISLAYY343 pKa = 9.58IDD345 pKa = 5.51DD346 pKa = 4.58IIVFTKK352 pKa = 9.22GTKK355 pKa = 9.64DD356 pKa = 3.32YY357 pKa = 10.9HH358 pKa = 6.46LKK360 pKa = 9.84QVARR364 pKa = 11.84VLIQLGNHH372 pKa = 5.35GVILSKK378 pKa = 10.84EE379 pKa = 3.81KK380 pKa = 10.98AKK382 pKa = 10.56IAFEE386 pKa = 4.34EE387 pKa = 4.55IEE389 pKa = 4.1FLGLKK394 pKa = 9.3ILKK397 pKa = 10.13NGFIEE402 pKa = 4.34PQKK405 pKa = 11.02HH406 pKa = 5.37LLEE409 pKa = 5.37KK410 pKa = 10.15IAEE413 pKa = 4.44FPDD416 pKa = 3.22QLQDD420 pKa = 2.92RR421 pKa = 11.84KK422 pKa = 10.55QIQKK426 pKa = 10.4FLGCLNYY433 pKa = 9.87IGEE436 pKa = 4.24KK437 pKa = 10.95GFFKK441 pKa = 10.68EE442 pKa = 3.76LAKK445 pKa = 10.27EE446 pKa = 4.02RR447 pKa = 11.84KK448 pKa = 9.07VLQKK452 pKa = 9.88MLSEE456 pKa = 4.03KK457 pKa = 10.8LPWKK461 pKa = 9.99WNDD464 pKa = 3.19LATLAVKK471 pKa = 10.36RR472 pKa = 11.84LKK474 pKa = 9.98QVCKK478 pKa = 10.14NLPRR482 pKa = 11.84LYY484 pKa = 10.09VAKK487 pKa = 10.36PSDD490 pKa = 4.22LLILTTDD497 pKa = 3.8ASDD500 pKa = 3.93TTWGAVLMAVPNAYY514 pKa = 9.9EE515 pKa = 4.01EE516 pKa = 4.31FLSFVTNNRR525 pKa = 11.84PSDD528 pKa = 3.38LKK530 pKa = 10.34TFLKK534 pKa = 10.46QKK536 pKa = 9.66EE537 pKa = 4.34GSSRR541 pKa = 11.84YY542 pKa = 9.41AEE544 pKa = 3.82KK545 pKa = 10.63SVPEE549 pKa = 4.6KK550 pKa = 10.64LNSMHH555 pKa = 7.21DD556 pKa = 3.36KK557 pKa = 10.82SHH559 pKa = 6.41GEE561 pKa = 3.77KK562 pKa = 10.49AEE564 pKa = 4.2KK565 pKa = 10.23SVSRR569 pKa = 11.84QTSQGRR575 pKa = 11.84SSSQSQQSYY584 pKa = 9.56IGSFMITKK592 pKa = 9.41WSSGTFKK599 pKa = 10.45PAEE602 pKa = 4.0EE603 pKa = 4.68NYY605 pKa = 9.09TVHH608 pKa = 6.7EE609 pKa = 4.49KK610 pKa = 10.86EE611 pKa = 4.29LLAFVNALKK620 pKa = 10.66KK621 pKa = 10.29FQIDD625 pKa = 3.5LRR627 pKa = 11.84PVKK630 pKa = 10.54FIFQTDD636 pKa = 3.66NSWVQGFLVNKK647 pKa = 10.34LKK649 pKa = 9.5EE650 pKa = 4.13TYY652 pKa = 10.3NRR654 pKa = 11.84GRR656 pKa = 11.84LARR659 pKa = 11.84WSMFIQQFDD668 pKa = 4.72FISLHH673 pKa = 5.62IAGKK677 pKa = 9.99EE678 pKa = 3.73NYY680 pKa = 9.95LADD683 pKa = 3.61TLTRR687 pKa = 11.84EE688 pKa = 4.4WKK690 pKa = 8.93TSSRR694 pKa = 3.78
Molecular weight: 80.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1896 |
320 |
694 |
474.0 |
54.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.325 ± 0.623 | 1.53 ± 0.427 |
5.222 ± 0.421 | 8.228 ± 0.875 |
3.956 ± 0.455 | 4.483 ± 0.127 |
1.319 ± 0.222 | 8.07 ± 0.897 |
9.968 ± 0.544 | 9.124 ± 0.901 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.741 ± 0.139 | 5.063 ± 0.49 |
4.061 ± 0.317 | 6.013 ± 0.212 |
5.38 ± 0.635 | 6.909 ± 0.567 |
4.747 ± 0.354 | 4.641 ± 0.611 |
0.844 ± 0.262 | 4.378 ± 0.565 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
