
Termite associated circular virus 4
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
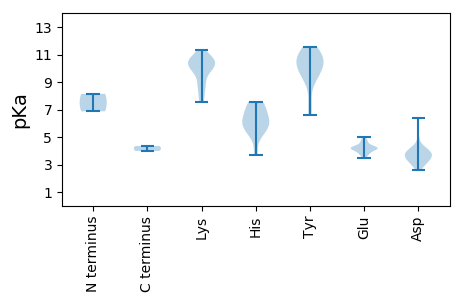
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
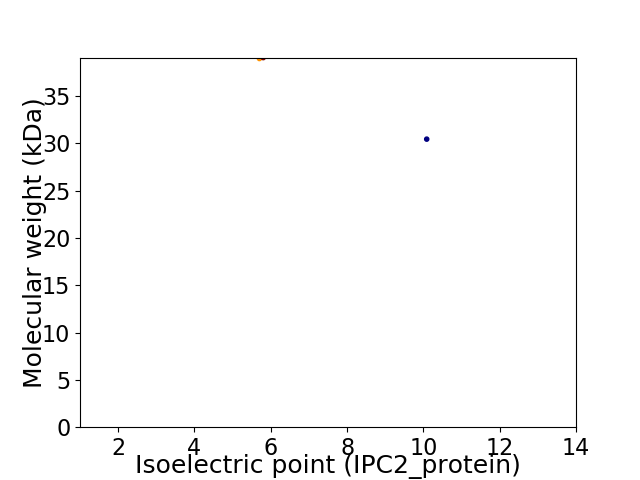
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1E315|A0A2P1E315_9VIRU Replication associated protein OS=Termite associated circular virus 4 OX=2108552 PE=3 SV=1
MM1 pKa = 8.14PEE3 pKa = 3.93FRR5 pKa = 11.84WQGKK9 pKa = 7.58YY10 pKa = 10.71LFLTYY15 pKa = 9.76PQSDD19 pKa = 4.02FNLDD23 pKa = 3.63GAVHH27 pKa = 7.3RR28 pKa = 11.84LRR30 pKa = 11.84EE31 pKa = 3.83QGGRR35 pKa = 11.84HH36 pKa = 3.73VTFVRR41 pKa = 11.84VCSEE45 pKa = 3.61QHH47 pKa = 5.52QSGEE51 pKa = 4.16LHH53 pKa = 6.13RR54 pKa = 11.84HH55 pKa = 5.41ALLCFGKK62 pKa = 10.23RR63 pKa = 11.84VSFTDD68 pKa = 3.07QRR70 pKa = 11.84RR71 pKa = 11.84FDD73 pKa = 3.28IDD75 pKa = 2.99GRR77 pKa = 11.84HH78 pKa = 6.06PKK80 pKa = 8.67WEE82 pKa = 4.16RR83 pKa = 11.84YY84 pKa = 6.57TRR86 pKa = 11.84SPAKK90 pKa = 9.72AAEE93 pKa = 4.28YY94 pKa = 9.38IAKK97 pKa = 10.08EE98 pKa = 4.1GEE100 pKa = 4.17YY101 pKa = 10.33VDD103 pKa = 3.76WGEE106 pKa = 4.31TPDD109 pKa = 4.94FSTPEE114 pKa = 3.91CRR116 pKa = 11.84EE117 pKa = 3.84SRR119 pKa = 11.84NEE121 pKa = 3.46LWGRR125 pKa = 11.84LLDD128 pKa = 3.93EE129 pKa = 4.53ATTPEE134 pKa = 4.2SFMSLVRR141 pKa = 11.84QFAPYY146 pKa = 10.76DD147 pKa = 3.2FATRR151 pKa = 11.84YY152 pKa = 8.74QALHH156 pKa = 5.69TMATSVFHH164 pKa = 6.81RR165 pKa = 11.84RR166 pKa = 11.84SPHH169 pKa = 5.15QSPHH173 pKa = 7.56DD174 pKa = 3.77MVEE177 pKa = 4.2DD178 pKa = 4.19FQLPDD183 pKa = 3.61TIEE186 pKa = 3.72RR187 pKa = 11.84WLEE190 pKa = 3.79EE191 pKa = 4.14EE192 pKa = 4.16FDD194 pKa = 3.66QEE196 pKa = 4.18NRR198 pKa = 11.84PIRR201 pKa = 11.84PKK203 pKa = 10.75SLLIVGDD210 pKa = 3.75SRR212 pKa = 11.84SGKK215 pKa = 8.7TSFARR220 pKa = 11.84CLGHH224 pKa = 7.21HH225 pKa = 7.15IYY227 pKa = 10.44FQGLFNLDD235 pKa = 3.58KK236 pKa = 10.7FDD238 pKa = 4.16PQADD242 pKa = 3.72YY243 pKa = 11.54AVFDD247 pKa = 3.85DD248 pKa = 6.37CKK250 pKa = 10.72IKK252 pKa = 10.14WISNHH257 pKa = 6.18KK258 pKa = 9.94SWLGCGGEE266 pKa = 4.7FEE268 pKa = 4.99ATDD271 pKa = 3.3KK272 pKa = 11.32YY273 pKa = 9.54RR274 pKa = 11.84TKK276 pKa = 10.36RR277 pKa = 11.84TLSWSGKK284 pKa = 9.53PSIICCNRR292 pKa = 11.84GSTWDD297 pKa = 3.2WRR299 pKa = 11.84YY300 pKa = 10.53SEE302 pKa = 4.54EE303 pKa = 4.3WKK305 pKa = 10.66DD306 pKa = 3.55DD307 pKa = 4.15PGWFEE312 pKa = 4.81ANVVVCEE319 pKa = 4.72LGPNQQLWEE328 pKa = 4.28VPNN331 pKa = 3.99
MM1 pKa = 8.14PEE3 pKa = 3.93FRR5 pKa = 11.84WQGKK9 pKa = 7.58YY10 pKa = 10.71LFLTYY15 pKa = 9.76PQSDD19 pKa = 4.02FNLDD23 pKa = 3.63GAVHH27 pKa = 7.3RR28 pKa = 11.84LRR30 pKa = 11.84EE31 pKa = 3.83QGGRR35 pKa = 11.84HH36 pKa = 3.73VTFVRR41 pKa = 11.84VCSEE45 pKa = 3.61QHH47 pKa = 5.52QSGEE51 pKa = 4.16LHH53 pKa = 6.13RR54 pKa = 11.84HH55 pKa = 5.41ALLCFGKK62 pKa = 10.23RR63 pKa = 11.84VSFTDD68 pKa = 3.07QRR70 pKa = 11.84RR71 pKa = 11.84FDD73 pKa = 3.28IDD75 pKa = 2.99GRR77 pKa = 11.84HH78 pKa = 6.06PKK80 pKa = 8.67WEE82 pKa = 4.16RR83 pKa = 11.84YY84 pKa = 6.57TRR86 pKa = 11.84SPAKK90 pKa = 9.72AAEE93 pKa = 4.28YY94 pKa = 9.38IAKK97 pKa = 10.08EE98 pKa = 4.1GEE100 pKa = 4.17YY101 pKa = 10.33VDD103 pKa = 3.76WGEE106 pKa = 4.31TPDD109 pKa = 4.94FSTPEE114 pKa = 3.91CRR116 pKa = 11.84EE117 pKa = 3.84SRR119 pKa = 11.84NEE121 pKa = 3.46LWGRR125 pKa = 11.84LLDD128 pKa = 3.93EE129 pKa = 4.53ATTPEE134 pKa = 4.2SFMSLVRR141 pKa = 11.84QFAPYY146 pKa = 10.76DD147 pKa = 3.2FATRR151 pKa = 11.84YY152 pKa = 8.74QALHH156 pKa = 5.69TMATSVFHH164 pKa = 6.81RR165 pKa = 11.84RR166 pKa = 11.84SPHH169 pKa = 5.15QSPHH173 pKa = 7.56DD174 pKa = 3.77MVEE177 pKa = 4.2DD178 pKa = 4.19FQLPDD183 pKa = 3.61TIEE186 pKa = 3.72RR187 pKa = 11.84WLEE190 pKa = 3.79EE191 pKa = 4.14EE192 pKa = 4.16FDD194 pKa = 3.66QEE196 pKa = 4.18NRR198 pKa = 11.84PIRR201 pKa = 11.84PKK203 pKa = 10.75SLLIVGDD210 pKa = 3.75SRR212 pKa = 11.84SGKK215 pKa = 8.7TSFARR220 pKa = 11.84CLGHH224 pKa = 7.21HH225 pKa = 7.15IYY227 pKa = 10.44FQGLFNLDD235 pKa = 3.58KK236 pKa = 10.7FDD238 pKa = 4.16PQADD242 pKa = 3.72YY243 pKa = 11.54AVFDD247 pKa = 3.85DD248 pKa = 6.37CKK250 pKa = 10.72IKK252 pKa = 10.14WISNHH257 pKa = 6.18KK258 pKa = 9.94SWLGCGGEE266 pKa = 4.7FEE268 pKa = 4.99ATDD271 pKa = 3.3KK272 pKa = 11.32YY273 pKa = 9.54RR274 pKa = 11.84TKK276 pKa = 10.36RR277 pKa = 11.84TLSWSGKK284 pKa = 9.53PSIICCNRR292 pKa = 11.84GSTWDD297 pKa = 3.2WRR299 pKa = 11.84YY300 pKa = 10.53SEE302 pKa = 4.54EE303 pKa = 4.3WKK305 pKa = 10.66DD306 pKa = 3.55DD307 pKa = 4.15PGWFEE312 pKa = 4.81ANVVVCEE319 pKa = 4.72LGPNQQLWEE328 pKa = 4.28VPNN331 pKa = 3.99
Molecular weight: 38.96 kDa
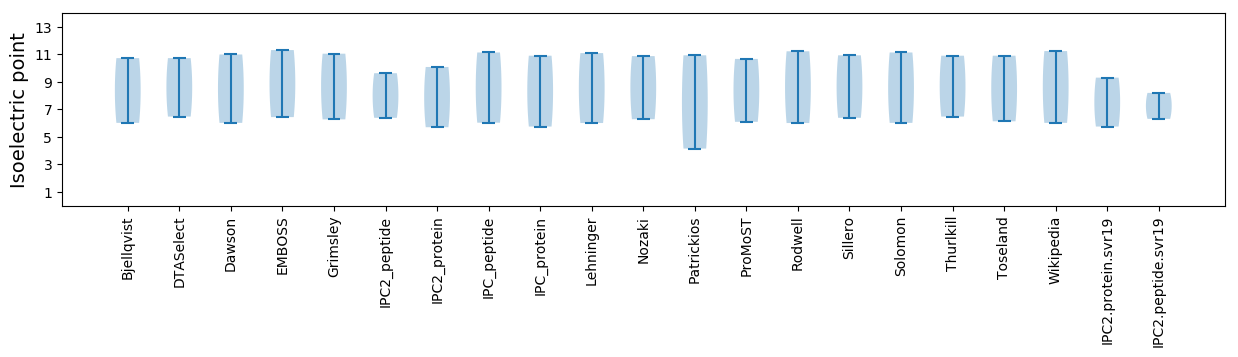
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1E315|A0A2P1E315_9VIRU Replication associated protein OS=Termite associated circular virus 4 OX=2108552 PE=3 SV=1
MM1 pKa = 6.87PQRR4 pKa = 11.84RR5 pKa = 11.84ARR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84TARR13 pKa = 11.84KK14 pKa = 8.77GRR16 pKa = 11.84KK17 pKa = 7.9STTTRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 8.72QPLVKK29 pKa = 10.42VMRR32 pKa = 11.84RR33 pKa = 11.84VARR36 pKa = 11.84QQINRR41 pKa = 11.84AVEE44 pKa = 4.19TKK46 pKa = 10.13RR47 pKa = 11.84FAIINEE53 pKa = 4.31SWSSTFIDD61 pKa = 4.24FATRR65 pKa = 11.84NTTWKK70 pKa = 10.08YY71 pKa = 11.28KK72 pKa = 10.85NLFSQLGTGNTSWSIVGNEE91 pKa = 4.17ILNPMVKK98 pKa = 9.43IKK100 pKa = 10.69GRR102 pKa = 11.84IWVDD106 pKa = 2.79WNANRR111 pKa = 11.84QINTVVGPQDD121 pKa = 3.52VQVTVYY127 pKa = 10.4LIAANEE133 pKa = 4.07QLSTTAPSPLLASSDD148 pKa = 3.43YY149 pKa = 10.74WFYY152 pKa = 11.36QPDD155 pKa = 3.99GQNPTLNGNNVKK167 pKa = 10.42VLGRR171 pKa = 11.84TSKK174 pKa = 10.52KK175 pKa = 10.06IKK177 pKa = 10.09AQLPIAAATTSTFSGTSTTRR197 pKa = 11.84FQMSYY202 pKa = 7.46RR203 pKa = 11.84WKK205 pKa = 10.64RR206 pKa = 11.84KK207 pKa = 7.73LTYY210 pKa = 9.94EE211 pKa = 4.21DD212 pKa = 4.09VVGNPPSVGGPTRR225 pKa = 11.84DD226 pKa = 3.04VYY228 pKa = 11.34LRR230 pKa = 11.84GWNYY234 pKa = 9.52YY235 pKa = 9.59VLLGVHH241 pKa = 6.35VPQTHH246 pKa = 6.16NGSLVGTSTNVSMDD260 pKa = 2.59TFMYY264 pKa = 10.82YY265 pKa = 10.34KK266 pKa = 10.69DD267 pKa = 3.5PP268 pKa = 4.35
MM1 pKa = 6.87PQRR4 pKa = 11.84RR5 pKa = 11.84ARR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84TARR13 pKa = 11.84KK14 pKa = 8.77GRR16 pKa = 11.84KK17 pKa = 7.9STTTRR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 8.72QPLVKK29 pKa = 10.42VMRR32 pKa = 11.84RR33 pKa = 11.84VARR36 pKa = 11.84QQINRR41 pKa = 11.84AVEE44 pKa = 4.19TKK46 pKa = 10.13RR47 pKa = 11.84FAIINEE53 pKa = 4.31SWSSTFIDD61 pKa = 4.24FATRR65 pKa = 11.84NTTWKK70 pKa = 10.08YY71 pKa = 11.28KK72 pKa = 10.85NLFSQLGTGNTSWSIVGNEE91 pKa = 4.17ILNPMVKK98 pKa = 9.43IKK100 pKa = 10.69GRR102 pKa = 11.84IWVDD106 pKa = 2.79WNANRR111 pKa = 11.84QINTVVGPQDD121 pKa = 3.52VQVTVYY127 pKa = 10.4LIAANEE133 pKa = 4.07QLSTTAPSPLLASSDD148 pKa = 3.43YY149 pKa = 10.74WFYY152 pKa = 11.36QPDD155 pKa = 3.99GQNPTLNGNNVKK167 pKa = 10.42VLGRR171 pKa = 11.84TSKK174 pKa = 10.52KK175 pKa = 10.06IKK177 pKa = 10.09AQLPIAAATTSTFSGTSTTRR197 pKa = 11.84FQMSYY202 pKa = 7.46RR203 pKa = 11.84WKK205 pKa = 10.64RR206 pKa = 11.84KK207 pKa = 7.73LTYY210 pKa = 9.94EE211 pKa = 4.21DD212 pKa = 4.09VVGNPPSVGGPTRR225 pKa = 11.84DD226 pKa = 3.04VYY228 pKa = 11.34LRR230 pKa = 11.84GWNYY234 pKa = 9.52YY235 pKa = 9.59VLLGVHH241 pKa = 6.35VPQTHH246 pKa = 6.16NGSLVGTSTNVSMDD260 pKa = 2.59TFMYY264 pKa = 10.82YY265 pKa = 10.34KK266 pKa = 10.69DD267 pKa = 3.5PP268 pKa = 4.35
Molecular weight: 30.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
599 |
268 |
331 |
299.5 |
34.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.175 ± 0.273 | 1.503 ± 0.973 |
5.342 ± 1.285 | 5.342 ± 2.251 |
4.841 ± 1.202 | 6.344 ± 0.0 |
2.504 ± 1.138 | 3.673 ± 0.521 |
5.175 ± 0.515 | 6.511 ± 0.35 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.669 ± 0.369 | 4.674 ± 1.564 |
5.342 ± 0.077 | 4.841 ± 0.248 |
8.514 ± 0.044 | 7.179 ± 0.184 |
7.846 ± 2.167 | 6.344 ± 1.449 |
3.506 ± 0.337 | 3.673 ± 0.279 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
