
CRESS virus sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
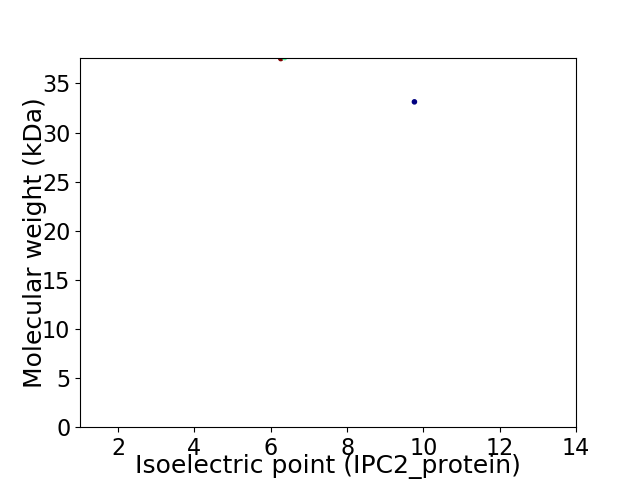
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A385E6C8|A0A385E6C8_9VIRU Putative capsid protein OS=CRESS virus sp. OX=2202563 PE=4 SV=1
MM1 pKa = 8.09KK2 pKa = 9.82IRR4 pKa = 11.84CMCGTLNNYY13 pKa = 7.45TVAGLAKK20 pKa = 10.65VKK22 pKa = 10.76DD23 pKa = 4.09FLEE26 pKa = 4.09NHH28 pKa = 5.77CTYY31 pKa = 10.74GVYY34 pKa = 10.57GEE36 pKa = 4.59EE37 pKa = 4.39VGEE40 pKa = 4.32SGTPHH45 pKa = 5.7LQIYY49 pKa = 10.18FEE51 pKa = 5.07MKK53 pKa = 9.03TQMTYY58 pKa = 10.73SAVNKK63 pKa = 10.39KK64 pKa = 10.56LGFKK68 pKa = 10.47KK69 pKa = 10.27GGEE72 pKa = 4.21WIDD75 pKa = 3.21LSTRR79 pKa = 11.84HH80 pKa = 5.47GTPEE84 pKa = 3.56QAAGYY89 pKa = 8.2CKK91 pKa = 10.26KK92 pKa = 10.79GNEE95 pKa = 4.21TEE97 pKa = 4.19EE98 pKa = 4.49VYY100 pKa = 10.81PDD102 pKa = 3.12GWTDD106 pKa = 3.41GAAWWFTRR114 pKa = 11.84PAASWIGDD122 pKa = 3.3EE123 pKa = 4.66WGTISNQGEE132 pKa = 4.18RR133 pKa = 11.84KK134 pKa = 9.55DD135 pKa = 3.7LKK137 pKa = 10.85RR138 pKa = 11.84KK139 pKa = 8.65IDD141 pKa = 3.71EE142 pKa = 4.69VISGDD147 pKa = 3.6TTPDD151 pKa = 3.83EE152 pKa = 4.81ICVEE156 pKa = 4.13MPVFYY161 pKa = 10.26HH162 pKa = 5.88QYY164 pKa = 10.74GRR166 pKa = 11.84TLEE169 pKa = 4.23KK170 pKa = 10.06AWCIAMRR177 pKa = 11.84KK178 pKa = 7.9KK179 pKa = 9.63WRR181 pKa = 11.84TEE183 pKa = 3.3MTTCDD188 pKa = 3.89WLWGEE193 pKa = 4.18TGVGKK198 pKa = 8.25SHH200 pKa = 7.21KK201 pKa = 10.15AFEE204 pKa = 5.02GYY206 pKa = 10.01SPEE209 pKa = 3.83THH211 pKa = 6.13YY212 pKa = 11.24VWNLAEE218 pKa = 4.59EE219 pKa = 4.54FQCGYY224 pKa = 9.79RR225 pKa = 11.84QQDD228 pKa = 3.38TVIINEE234 pKa = 3.58FRR236 pKa = 11.84GQIKK240 pKa = 10.47YY241 pKa = 10.61SEE243 pKa = 4.82LLTLIDD249 pKa = 4.49KK250 pKa = 9.54WPHH253 pKa = 5.12CIKK256 pKa = 10.72RR257 pKa = 11.84KK258 pKa = 10.16GGEE261 pKa = 3.74QLPFVSKK268 pKa = 10.52HH269 pKa = 6.23IIITSCKK276 pKa = 9.26PPDD279 pKa = 4.33EE280 pKa = 4.53IYY282 pKa = 10.4PRR284 pKa = 11.84QDD286 pKa = 2.9EE287 pKa = 4.37KK288 pKa = 11.71DD289 pKa = 3.39SLRR292 pKa = 11.84QLLRR296 pKa = 11.84RR297 pKa = 11.84IKK299 pKa = 10.35VIEE302 pKa = 4.19VEE304 pKa = 4.15KK305 pKa = 10.78QPPVWGRR312 pKa = 11.84APPYY316 pKa = 8.38PPPMTACGGCGG327 pKa = 2.94
MM1 pKa = 8.09KK2 pKa = 9.82IRR4 pKa = 11.84CMCGTLNNYY13 pKa = 7.45TVAGLAKK20 pKa = 10.65VKK22 pKa = 10.76DD23 pKa = 4.09FLEE26 pKa = 4.09NHH28 pKa = 5.77CTYY31 pKa = 10.74GVYY34 pKa = 10.57GEE36 pKa = 4.59EE37 pKa = 4.39VGEE40 pKa = 4.32SGTPHH45 pKa = 5.7LQIYY49 pKa = 10.18FEE51 pKa = 5.07MKK53 pKa = 9.03TQMTYY58 pKa = 10.73SAVNKK63 pKa = 10.39KK64 pKa = 10.56LGFKK68 pKa = 10.47KK69 pKa = 10.27GGEE72 pKa = 4.21WIDD75 pKa = 3.21LSTRR79 pKa = 11.84HH80 pKa = 5.47GTPEE84 pKa = 3.56QAAGYY89 pKa = 8.2CKK91 pKa = 10.26KK92 pKa = 10.79GNEE95 pKa = 4.21TEE97 pKa = 4.19EE98 pKa = 4.49VYY100 pKa = 10.81PDD102 pKa = 3.12GWTDD106 pKa = 3.41GAAWWFTRR114 pKa = 11.84PAASWIGDD122 pKa = 3.3EE123 pKa = 4.66WGTISNQGEE132 pKa = 4.18RR133 pKa = 11.84KK134 pKa = 9.55DD135 pKa = 3.7LKK137 pKa = 10.85RR138 pKa = 11.84KK139 pKa = 8.65IDD141 pKa = 3.71EE142 pKa = 4.69VISGDD147 pKa = 3.6TTPDD151 pKa = 3.83EE152 pKa = 4.81ICVEE156 pKa = 4.13MPVFYY161 pKa = 10.26HH162 pKa = 5.88QYY164 pKa = 10.74GRR166 pKa = 11.84TLEE169 pKa = 4.23KK170 pKa = 10.06AWCIAMRR177 pKa = 11.84KK178 pKa = 7.9KK179 pKa = 9.63WRR181 pKa = 11.84TEE183 pKa = 3.3MTTCDD188 pKa = 3.89WLWGEE193 pKa = 4.18TGVGKK198 pKa = 8.25SHH200 pKa = 7.21KK201 pKa = 10.15AFEE204 pKa = 5.02GYY206 pKa = 10.01SPEE209 pKa = 3.83THH211 pKa = 6.13YY212 pKa = 11.24VWNLAEE218 pKa = 4.59EE219 pKa = 4.54FQCGYY224 pKa = 9.79RR225 pKa = 11.84QQDD228 pKa = 3.38TVIINEE234 pKa = 3.58FRR236 pKa = 11.84GQIKK240 pKa = 10.47YY241 pKa = 10.61SEE243 pKa = 4.82LLTLIDD249 pKa = 4.49KK250 pKa = 9.54WPHH253 pKa = 5.12CIKK256 pKa = 10.72RR257 pKa = 11.84KK258 pKa = 10.16GGEE261 pKa = 3.74QLPFVSKK268 pKa = 10.52HH269 pKa = 6.23IIITSCKK276 pKa = 9.26PPDD279 pKa = 4.33EE280 pKa = 4.53IYY282 pKa = 10.4PRR284 pKa = 11.84QDD286 pKa = 2.9EE287 pKa = 4.37KK288 pKa = 11.71DD289 pKa = 3.39SLRR292 pKa = 11.84QLLRR296 pKa = 11.84RR297 pKa = 11.84IKK299 pKa = 10.35VIEE302 pKa = 4.19VEE304 pKa = 4.15KK305 pKa = 10.78QPPVWGRR312 pKa = 11.84APPYY316 pKa = 8.38PPPMTACGGCGG327 pKa = 2.94
Molecular weight: 37.53 kDa
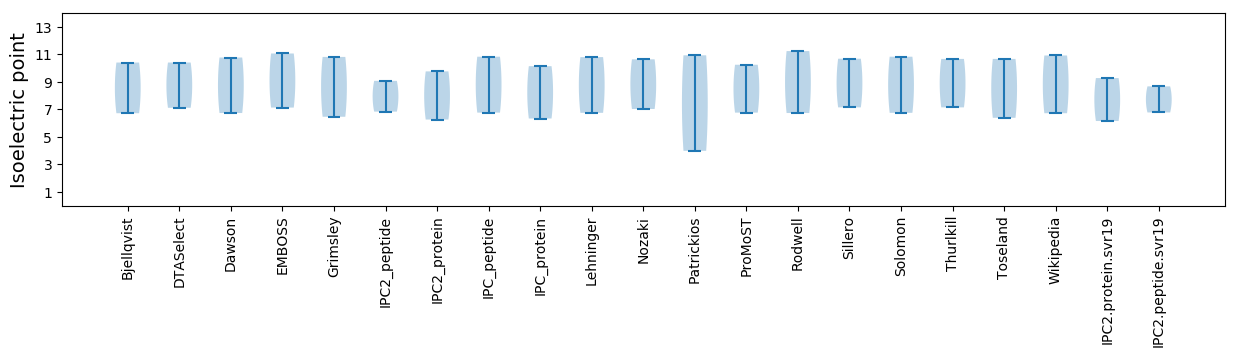
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A385E6C8|A0A385E6C8_9VIRU Putative capsid protein OS=CRESS virus sp. OX=2202563 PE=4 SV=1
MM1 pKa = 7.35LRR3 pKa = 11.84KK4 pKa = 9.93KK5 pKa = 10.44ISPKK9 pKa = 9.32FKK11 pKa = 9.55KK12 pKa = 9.77RR13 pKa = 11.84NMARR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 5.02YY20 pKa = 10.3GKK22 pKa = 10.14KK23 pKa = 9.74RR24 pKa = 11.84PRR26 pKa = 11.84KK27 pKa = 5.41TTSKK31 pKa = 10.48AAIMYY36 pKa = 7.42KK37 pKa = 10.14KK38 pKa = 10.0PSASNQQKK46 pKa = 10.16QIAALAVKK54 pKa = 9.84VNRR57 pKa = 11.84NQRR60 pKa = 11.84RR61 pKa = 11.84INQQRR66 pKa = 11.84YY67 pKa = 8.18LVQHH71 pKa = 3.99QTKK74 pKa = 9.95FEE76 pKa = 4.12EE77 pKa = 4.49QPVTRR82 pKa = 11.84PAGYY86 pKa = 10.46APYY89 pKa = 9.14TAWSLNNIPFMEE101 pKa = 4.53QIFGDD106 pKa = 3.8PEE108 pKa = 3.7EE109 pKa = 4.66AKK111 pKa = 10.53GGKK114 pKa = 7.94YY115 pKa = 8.49TGKK118 pKa = 10.64ALRR121 pKa = 11.84LDD123 pKa = 3.76FNIGIGKK130 pKa = 7.46STRR133 pKa = 11.84TTNFTAFIIRR143 pKa = 11.84PKK145 pKa = 6.97TQKK148 pKa = 9.93VVNEE152 pKa = 4.49CGISQNNVLSPTSVLPSPMIAGTDD176 pKa = 3.72FSYY179 pKa = 11.09EE180 pKa = 3.83GGLALINPKK189 pKa = 9.53RR190 pKa = 11.84WHH192 pKa = 6.5IDD194 pKa = 2.6KK195 pKa = 10.45MMKK198 pKa = 10.1LQIRR202 pKa = 11.84PQVAPFYY209 pKa = 10.4QGTPPTPTNVVTQDD223 pKa = 2.65NRR225 pKa = 11.84VRR227 pKa = 11.84RR228 pKa = 11.84SCTLKK233 pKa = 10.36NPMYY237 pKa = 10.23INSRR241 pKa = 11.84TGDD244 pKa = 3.33WKK246 pKa = 8.64TTTDD250 pKa = 3.23PWEE253 pKa = 4.18LPARR257 pKa = 11.84QRR259 pKa = 11.84SFLVLFDD266 pKa = 4.51DD267 pKa = 4.8TPATTGTSPGPTFEE281 pKa = 4.62GLVHH285 pKa = 6.06LTAHH289 pKa = 6.01TSEE292 pKa = 4.32
MM1 pKa = 7.35LRR3 pKa = 11.84KK4 pKa = 9.93KK5 pKa = 10.44ISPKK9 pKa = 9.32FKK11 pKa = 9.55KK12 pKa = 9.77RR13 pKa = 11.84NMARR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 5.02YY20 pKa = 10.3GKK22 pKa = 10.14KK23 pKa = 9.74RR24 pKa = 11.84PRR26 pKa = 11.84KK27 pKa = 5.41TTSKK31 pKa = 10.48AAIMYY36 pKa = 7.42KK37 pKa = 10.14KK38 pKa = 10.0PSASNQQKK46 pKa = 10.16QIAALAVKK54 pKa = 9.84VNRR57 pKa = 11.84NQRR60 pKa = 11.84RR61 pKa = 11.84INQQRR66 pKa = 11.84YY67 pKa = 8.18LVQHH71 pKa = 3.99QTKK74 pKa = 9.95FEE76 pKa = 4.12EE77 pKa = 4.49QPVTRR82 pKa = 11.84PAGYY86 pKa = 10.46APYY89 pKa = 9.14TAWSLNNIPFMEE101 pKa = 4.53QIFGDD106 pKa = 3.8PEE108 pKa = 3.7EE109 pKa = 4.66AKK111 pKa = 10.53GGKK114 pKa = 7.94YY115 pKa = 8.49TGKK118 pKa = 10.64ALRR121 pKa = 11.84LDD123 pKa = 3.76FNIGIGKK130 pKa = 7.46STRR133 pKa = 11.84TTNFTAFIIRR143 pKa = 11.84PKK145 pKa = 6.97TQKK148 pKa = 9.93VVNEE152 pKa = 4.49CGISQNNVLSPTSVLPSPMIAGTDD176 pKa = 3.72FSYY179 pKa = 11.09EE180 pKa = 3.83GGLALINPKK189 pKa = 9.53RR190 pKa = 11.84WHH192 pKa = 6.5IDD194 pKa = 2.6KK195 pKa = 10.45MMKK198 pKa = 10.1LQIRR202 pKa = 11.84PQVAPFYY209 pKa = 10.4QGTPPTPTNVVTQDD223 pKa = 2.65NRR225 pKa = 11.84VRR227 pKa = 11.84RR228 pKa = 11.84SCTLKK233 pKa = 10.36NPMYY237 pKa = 10.23INSRR241 pKa = 11.84TGDD244 pKa = 3.33WKK246 pKa = 8.64TTTDD250 pKa = 3.23PWEE253 pKa = 4.18LPARR257 pKa = 11.84QRR259 pKa = 11.84SFLVLFDD266 pKa = 4.51DD267 pKa = 4.8TPATTGTSPGPTFEE281 pKa = 4.62GLVHH285 pKa = 6.06LTAHH289 pKa = 6.01TSEE292 pKa = 4.32
Molecular weight: 33.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
619 |
292 |
327 |
309.5 |
35.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.493 ± 0.658 | 2.262 ± 1.023 |
3.877 ± 0.516 | 6.3 ± 1.866 |
3.393 ± 0.465 | 7.754 ± 1.254 |
2.1 ± 0.252 | 5.816 ± 0.218 |
8.401 ± 0.104 | 5.493 ± 0.214 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.585 ± 0.101 | 4.039 ± 1.157 |
6.947 ± 0.826 | 4.847 ± 0.633 |
6.139 ± 0.905 | 4.523 ± 0.62 |
8.562 ± 0.889 | 4.847 ± 0.034 |
2.746 ± 0.893 | 3.877 ± 0.516 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
