
Capybara microvirus Cap1_SP_87
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.04
Get precalculated fractions of proteins
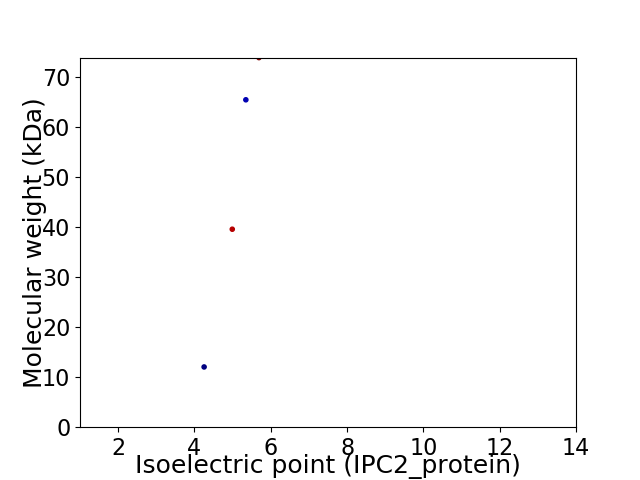
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W529|A0A4P8W529_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_87 OX=2584796 PE=3 SV=1
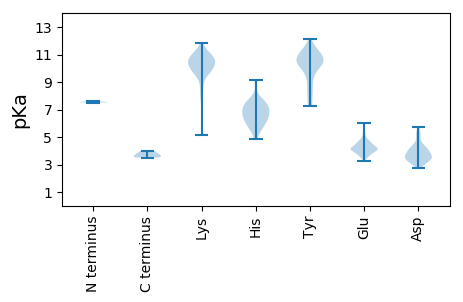
MM1 pKa = 7.54KK2 pKa = 9.58VTKK5 pKa = 10.23LLYY8 pKa = 10.56LEE10 pKa = 4.3NPRR13 pKa = 11.84YY14 pKa = 9.56ASPIIPDD21 pKa = 3.12QVMEE25 pKa = 4.46KK26 pKa = 10.44EE27 pKa = 4.1YY28 pKa = 10.93TEE30 pKa = 4.88DD31 pKa = 3.81GNVVQVWKK39 pKa = 10.83VKK41 pKa = 10.81SNYY44 pKa = 9.28EE45 pKa = 4.05DD46 pKa = 5.33LIEE49 pKa = 4.57LHH51 pKa = 7.01GDD53 pKa = 3.33ANNWSIDD60 pKa = 3.43ALMKK64 pKa = 10.8AGIDD68 pKa = 3.62PASTSINTGKK78 pKa = 10.27NSRR81 pKa = 11.84LDD83 pKa = 3.67GYY85 pKa = 11.49NDD87 pKa = 3.46LGKK90 pKa = 10.62FIDD93 pKa = 4.3GFEE96 pKa = 4.1VLTDD100 pKa = 3.93EE101 pKa = 6.04GDD103 pKa = 3.25IDD105 pKa = 4.26NNN107 pKa = 3.74
MM1 pKa = 7.54KK2 pKa = 9.58VTKK5 pKa = 10.23LLYY8 pKa = 10.56LEE10 pKa = 4.3NPRR13 pKa = 11.84YY14 pKa = 9.56ASPIIPDD21 pKa = 3.12QVMEE25 pKa = 4.46KK26 pKa = 10.44EE27 pKa = 4.1YY28 pKa = 10.93TEE30 pKa = 4.88DD31 pKa = 3.81GNVVQVWKK39 pKa = 10.83VKK41 pKa = 10.81SNYY44 pKa = 9.28EE45 pKa = 4.05DD46 pKa = 5.33LIEE49 pKa = 4.57LHH51 pKa = 7.01GDD53 pKa = 3.33ANNWSIDD60 pKa = 3.43ALMKK64 pKa = 10.8AGIDD68 pKa = 3.62PASTSINTGKK78 pKa = 10.27NSRR81 pKa = 11.84LDD83 pKa = 3.67GYY85 pKa = 11.49NDD87 pKa = 3.46LGKK90 pKa = 10.62FIDD93 pKa = 4.3GFEE96 pKa = 4.1VLTDD100 pKa = 3.93EE101 pKa = 6.04GDD103 pKa = 3.25IDD105 pKa = 4.26NNN107 pKa = 3.74
Molecular weight: 12.03 kDa
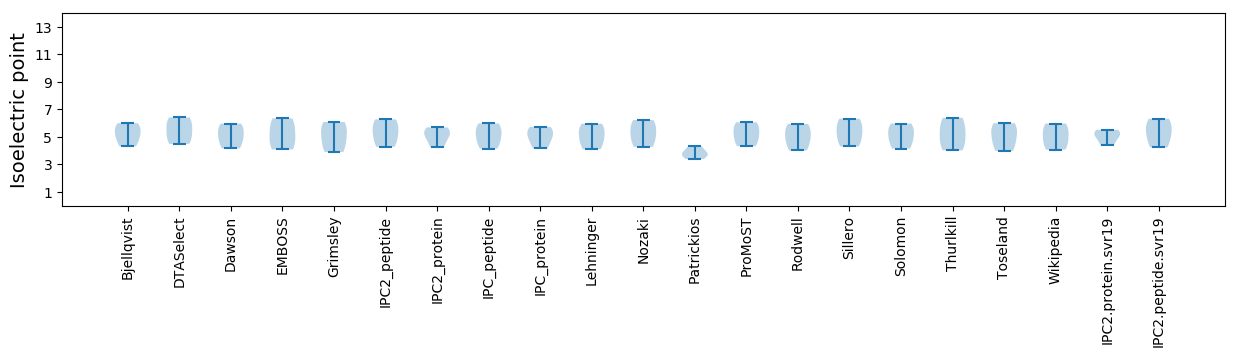
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7M9|A0A4P8W7M9_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_87 OX=2584796 PE=4 SV=1
MM1 pKa = 7.51FKK3 pKa = 10.0TLTVKK8 pKa = 9.25NTTHH12 pKa = 6.29YY13 pKa = 11.32VDD15 pKa = 4.36LSRR18 pKa = 11.84SKK20 pKa = 10.19ICNSFRR26 pKa = 11.84NPSAFQSEE34 pKa = 4.32EE35 pKa = 4.19LKK37 pKa = 10.34RR38 pKa = 11.84QSYY41 pKa = 7.25EE42 pKa = 3.28ARR44 pKa = 11.84LYY46 pKa = 10.96YY47 pKa = 10.25EE48 pKa = 4.72YY49 pKa = 11.0LDD51 pKa = 3.85CQRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 10.54CPVFFYY62 pKa = 10.7TLTFNNMHH70 pKa = 6.64LPHH73 pKa = 7.3YY74 pKa = 8.51EE75 pKa = 3.75GHH77 pKa = 5.8VCFDD81 pKa = 4.08NKK83 pKa = 10.57ILMQFMNSLRR93 pKa = 11.84KK94 pKa = 9.62RR95 pKa = 11.84LVRR98 pKa = 11.84DD99 pKa = 3.17YY100 pKa = 11.41GCNFKK105 pKa = 11.2YY106 pKa = 10.17FVSCEE111 pKa = 3.94MGEE114 pKa = 4.33GAGTRR119 pKa = 11.84KK120 pKa = 9.85EE121 pKa = 4.22YY122 pKa = 10.78EE123 pKa = 3.87NPHH126 pKa = 4.88YY127 pKa = 10.44HH128 pKa = 6.38VLFFTYY134 pKa = 10.43AEE136 pKa = 4.36PDD138 pKa = 3.33HH139 pKa = 6.72YY140 pKa = 10.46FVRR143 pKa = 11.84PTVYY147 pKa = 10.4DD148 pKa = 3.98FKK150 pKa = 11.48KK151 pKa = 10.35LVRR154 pKa = 11.84LYY156 pKa = 10.62WMGQEE161 pKa = 3.64KK162 pKa = 10.43VGYY165 pKa = 9.73INYY168 pKa = 7.73KK169 pKa = 5.18TAKK172 pKa = 9.64YY173 pKa = 8.76GISMEE178 pKa = 4.23GDD180 pKa = 2.98HH181 pKa = 7.07CGLVLNFNACTYY193 pKa = 8.1VAKK196 pKa = 10.28YY197 pKa = 9.37VCKK200 pKa = 10.14DD201 pKa = 3.05TTKK204 pKa = 9.96WKK206 pKa = 10.59NIEE209 pKa = 4.16STNLSDD215 pKa = 4.41RR216 pKa = 11.84LYY218 pKa = 11.34SLVDD222 pKa = 3.45DD223 pKa = 4.5EE224 pKa = 5.25QNSKK228 pKa = 11.13EE229 pKa = 3.93NMVEE233 pKa = 4.09FFKK236 pKa = 11.45SEE238 pKa = 4.12FNPLLEE244 pKa = 4.68GKK246 pKa = 9.75SPQEE250 pKa = 3.74WYY252 pKa = 10.39DD253 pKa = 3.59EE254 pKa = 4.4YY255 pKa = 11.12IEE257 pKa = 4.56FLDD260 pKa = 4.9TIPFTYY266 pKa = 10.52DD267 pKa = 3.48DD268 pKa = 5.33LDD270 pKa = 5.4SEE272 pKa = 5.25DD273 pKa = 5.75DD274 pKa = 4.13YY275 pKa = 12.1DD276 pKa = 5.31DD277 pKa = 4.03EE278 pKa = 4.59EE279 pKa = 4.75NYY281 pKa = 10.23RR282 pKa = 11.84EE283 pKa = 4.08YY284 pKa = 11.29LDD286 pKa = 4.4KK287 pKa = 10.79KK288 pKa = 9.53AKK290 pKa = 9.84KK291 pKa = 9.81RR292 pKa = 11.84VILDD296 pKa = 3.52HH297 pKa = 5.82IKK299 pKa = 10.34KK300 pKa = 10.42YY301 pKa = 10.39KK302 pKa = 10.21RR303 pKa = 11.84CDD305 pKa = 4.26DD306 pKa = 3.85ILSHH310 pKa = 7.29LDD312 pKa = 3.51VIEE315 pKa = 4.61TILYY319 pKa = 9.94DD320 pKa = 3.72FNLYY324 pKa = 10.76DD325 pKa = 3.36NEE327 pKa = 4.47YY328 pKa = 11.21YY329 pKa = 10.87SFCCYY334 pKa = 10.61LSDD337 pKa = 5.54DD338 pKa = 4.4KK339 pKa = 11.81VSEE342 pKa = 4.04QKK344 pKa = 10.58KK345 pKa = 8.14EE346 pKa = 3.79FRR348 pKa = 11.84SRR350 pKa = 11.84YY351 pKa = 7.62CPKK354 pKa = 10.54VMISQGVGKK363 pKa = 10.43YY364 pKa = 10.37ALDD367 pKa = 5.16FINPDD372 pKa = 2.91HH373 pKa = 7.35PYY375 pKa = 10.96VVIQTTKK382 pKa = 9.81GPQSRR387 pKa = 11.84PVSLYY392 pKa = 8.54YY393 pKa = 10.22YY394 pKa = 10.34RR395 pKa = 11.84KK396 pKa = 9.79IYY398 pKa = 10.52CDD400 pKa = 3.28TLKK403 pKa = 10.91DD404 pKa = 3.58EE405 pKa = 4.98KK406 pKa = 10.99GQNIYY411 pKa = 10.61VLNSRR416 pKa = 11.84GIDD419 pKa = 3.08MRR421 pKa = 11.84MRR423 pKa = 11.84NLSKK427 pKa = 10.66EE428 pKa = 4.06LTKK431 pKa = 11.13LIDD434 pKa = 3.56DD435 pKa = 3.88TRR437 pKa = 11.84KK438 pKa = 10.16VIQLFRR444 pKa = 11.84DD445 pKa = 3.56DD446 pKa = 3.65SYY448 pKa = 11.84KK449 pKa = 11.04AEE451 pKa = 4.56FYY453 pKa = 11.22ANQRR457 pKa = 11.84EE458 pKa = 4.66YY459 pKa = 11.34LFHH462 pKa = 7.58PLNKK466 pKa = 10.06YY467 pKa = 7.57NTSLGDD473 pKa = 3.0IWNDD477 pKa = 2.72IHH479 pKa = 9.14SDD481 pKa = 3.21EE482 pKa = 5.36DD483 pKa = 4.81FDD485 pKa = 4.12MKK487 pKa = 11.13LNNIEE492 pKa = 4.15NKK494 pKa = 10.42LYY496 pKa = 10.69DD497 pKa = 3.43YY498 pKa = 11.05AIYY501 pKa = 10.46KK502 pKa = 9.88KK503 pKa = 9.61IYY505 pKa = 7.31EE506 pKa = 4.12WRR508 pKa = 11.84SYY510 pKa = 10.32RR511 pKa = 11.84QDD513 pKa = 3.25GDD515 pKa = 4.07SVSVIDD521 pKa = 4.41PEE523 pKa = 4.08YY524 pKa = 10.83DD525 pKa = 3.49YY526 pKa = 11.86SCFLHH531 pKa = 7.1SPALDD536 pKa = 3.47CSYY539 pKa = 10.92LPNGFDD545 pKa = 5.46DD546 pKa = 4.5ISDD549 pKa = 4.16SEE551 pKa = 4.31TSKK554 pKa = 11.34YY555 pKa = 11.01NLYY558 pKa = 10.27CDD560 pKa = 3.7HH561 pKa = 7.49PRR563 pKa = 11.84FKK565 pKa = 11.16CNIGLFTLLDD575 pKa = 3.82CMVDD579 pKa = 3.34YY580 pKa = 11.37AFWQKK585 pKa = 11.33DD586 pKa = 2.97KK587 pKa = 11.32AKK589 pKa = 9.91RR590 pKa = 11.84RR591 pKa = 11.84RR592 pKa = 11.84YY593 pKa = 8.17EE594 pKa = 3.52EE595 pKa = 3.8RR596 pKa = 11.84NKK598 pKa = 9.71VRR600 pKa = 11.84KK601 pKa = 7.8QQIVEE606 pKa = 4.32KK607 pKa = 10.54ISTFFNN613 pKa = 3.59
MM1 pKa = 7.51FKK3 pKa = 10.0TLTVKK8 pKa = 9.25NTTHH12 pKa = 6.29YY13 pKa = 11.32VDD15 pKa = 4.36LSRR18 pKa = 11.84SKK20 pKa = 10.19ICNSFRR26 pKa = 11.84NPSAFQSEE34 pKa = 4.32EE35 pKa = 4.19LKK37 pKa = 10.34RR38 pKa = 11.84QSYY41 pKa = 7.25EE42 pKa = 3.28ARR44 pKa = 11.84LYY46 pKa = 10.96YY47 pKa = 10.25EE48 pKa = 4.72YY49 pKa = 11.0LDD51 pKa = 3.85CQRR54 pKa = 11.84RR55 pKa = 11.84KK56 pKa = 10.54CPVFFYY62 pKa = 10.7TLTFNNMHH70 pKa = 6.64LPHH73 pKa = 7.3YY74 pKa = 8.51EE75 pKa = 3.75GHH77 pKa = 5.8VCFDD81 pKa = 4.08NKK83 pKa = 10.57ILMQFMNSLRR93 pKa = 11.84KK94 pKa = 9.62RR95 pKa = 11.84LVRR98 pKa = 11.84DD99 pKa = 3.17YY100 pKa = 11.41GCNFKK105 pKa = 11.2YY106 pKa = 10.17FVSCEE111 pKa = 3.94MGEE114 pKa = 4.33GAGTRR119 pKa = 11.84KK120 pKa = 9.85EE121 pKa = 4.22YY122 pKa = 10.78EE123 pKa = 3.87NPHH126 pKa = 4.88YY127 pKa = 10.44HH128 pKa = 6.38VLFFTYY134 pKa = 10.43AEE136 pKa = 4.36PDD138 pKa = 3.33HH139 pKa = 6.72YY140 pKa = 10.46FVRR143 pKa = 11.84PTVYY147 pKa = 10.4DD148 pKa = 3.98FKK150 pKa = 11.48KK151 pKa = 10.35LVRR154 pKa = 11.84LYY156 pKa = 10.62WMGQEE161 pKa = 3.64KK162 pKa = 10.43VGYY165 pKa = 9.73INYY168 pKa = 7.73KK169 pKa = 5.18TAKK172 pKa = 9.64YY173 pKa = 8.76GISMEE178 pKa = 4.23GDD180 pKa = 2.98HH181 pKa = 7.07CGLVLNFNACTYY193 pKa = 8.1VAKK196 pKa = 10.28YY197 pKa = 9.37VCKK200 pKa = 10.14DD201 pKa = 3.05TTKK204 pKa = 9.96WKK206 pKa = 10.59NIEE209 pKa = 4.16STNLSDD215 pKa = 4.41RR216 pKa = 11.84LYY218 pKa = 11.34SLVDD222 pKa = 3.45DD223 pKa = 4.5EE224 pKa = 5.25QNSKK228 pKa = 11.13EE229 pKa = 3.93NMVEE233 pKa = 4.09FFKK236 pKa = 11.45SEE238 pKa = 4.12FNPLLEE244 pKa = 4.68GKK246 pKa = 9.75SPQEE250 pKa = 3.74WYY252 pKa = 10.39DD253 pKa = 3.59EE254 pKa = 4.4YY255 pKa = 11.12IEE257 pKa = 4.56FLDD260 pKa = 4.9TIPFTYY266 pKa = 10.52DD267 pKa = 3.48DD268 pKa = 5.33LDD270 pKa = 5.4SEE272 pKa = 5.25DD273 pKa = 5.75DD274 pKa = 4.13YY275 pKa = 12.1DD276 pKa = 5.31DD277 pKa = 4.03EE278 pKa = 4.59EE279 pKa = 4.75NYY281 pKa = 10.23RR282 pKa = 11.84EE283 pKa = 4.08YY284 pKa = 11.29LDD286 pKa = 4.4KK287 pKa = 10.79KK288 pKa = 9.53AKK290 pKa = 9.84KK291 pKa = 9.81RR292 pKa = 11.84VILDD296 pKa = 3.52HH297 pKa = 5.82IKK299 pKa = 10.34KK300 pKa = 10.42YY301 pKa = 10.39KK302 pKa = 10.21RR303 pKa = 11.84CDD305 pKa = 4.26DD306 pKa = 3.85ILSHH310 pKa = 7.29LDD312 pKa = 3.51VIEE315 pKa = 4.61TILYY319 pKa = 9.94DD320 pKa = 3.72FNLYY324 pKa = 10.76DD325 pKa = 3.36NEE327 pKa = 4.47YY328 pKa = 11.21YY329 pKa = 10.87SFCCYY334 pKa = 10.61LSDD337 pKa = 5.54DD338 pKa = 4.4KK339 pKa = 11.81VSEE342 pKa = 4.04QKK344 pKa = 10.58KK345 pKa = 8.14EE346 pKa = 3.79FRR348 pKa = 11.84SRR350 pKa = 11.84YY351 pKa = 7.62CPKK354 pKa = 10.54VMISQGVGKK363 pKa = 10.43YY364 pKa = 10.37ALDD367 pKa = 5.16FINPDD372 pKa = 2.91HH373 pKa = 7.35PYY375 pKa = 10.96VVIQTTKK382 pKa = 9.81GPQSRR387 pKa = 11.84PVSLYY392 pKa = 8.54YY393 pKa = 10.22YY394 pKa = 10.34RR395 pKa = 11.84KK396 pKa = 9.79IYY398 pKa = 10.52CDD400 pKa = 3.28TLKK403 pKa = 10.91DD404 pKa = 3.58EE405 pKa = 4.98KK406 pKa = 10.99GQNIYY411 pKa = 10.61VLNSRR416 pKa = 11.84GIDD419 pKa = 3.08MRR421 pKa = 11.84MRR423 pKa = 11.84NLSKK427 pKa = 10.66EE428 pKa = 4.06LTKK431 pKa = 11.13LIDD434 pKa = 3.56DD435 pKa = 3.88TRR437 pKa = 11.84KK438 pKa = 10.16VIQLFRR444 pKa = 11.84DD445 pKa = 3.56DD446 pKa = 3.65SYY448 pKa = 11.84KK449 pKa = 11.04AEE451 pKa = 4.56FYY453 pKa = 11.22ANQRR457 pKa = 11.84EE458 pKa = 4.66YY459 pKa = 11.34LFHH462 pKa = 7.58PLNKK466 pKa = 10.06YY467 pKa = 7.57NTSLGDD473 pKa = 3.0IWNDD477 pKa = 2.72IHH479 pKa = 9.14SDD481 pKa = 3.21EE482 pKa = 5.36DD483 pKa = 4.81FDD485 pKa = 4.12MKK487 pKa = 11.13LNNIEE492 pKa = 4.15NKK494 pKa = 10.42LYY496 pKa = 10.69DD497 pKa = 3.43YY498 pKa = 11.05AIYY501 pKa = 10.46KK502 pKa = 9.88KK503 pKa = 9.61IYY505 pKa = 7.31EE506 pKa = 4.12WRR508 pKa = 11.84SYY510 pKa = 10.32RR511 pKa = 11.84QDD513 pKa = 3.25GDD515 pKa = 4.07SVSVIDD521 pKa = 4.41PEE523 pKa = 4.08YY524 pKa = 10.83DD525 pKa = 3.49YY526 pKa = 11.86SCFLHH531 pKa = 7.1SPALDD536 pKa = 3.47CSYY539 pKa = 10.92LPNGFDD545 pKa = 5.46DD546 pKa = 4.5ISDD549 pKa = 4.16SEE551 pKa = 4.31TSKK554 pKa = 11.34YY555 pKa = 11.01NLYY558 pKa = 10.27CDD560 pKa = 3.7HH561 pKa = 7.49PRR563 pKa = 11.84FKK565 pKa = 11.16CNIGLFTLLDD575 pKa = 3.82CMVDD579 pKa = 3.34YY580 pKa = 11.37AFWQKK585 pKa = 11.33DD586 pKa = 2.97KK587 pKa = 11.32AKK589 pKa = 9.91RR590 pKa = 11.84RR591 pKa = 11.84RR592 pKa = 11.84YY593 pKa = 8.17EE594 pKa = 3.52EE595 pKa = 3.8RR596 pKa = 11.84NKK598 pKa = 9.71VRR600 pKa = 11.84KK601 pKa = 7.8QQIVEE606 pKa = 4.32KK607 pKa = 10.54ISTFFNN613 pKa = 3.59
Molecular weight: 73.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669 |
107 |
613 |
417.3 |
47.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.531 ± 2.0 | 1.558 ± 0.669 |
7.849 ± 0.824 | 5.632 ± 0.952 |
4.913 ± 0.638 | 5.692 ± 1.04 |
1.558 ± 0.529 | 5.153 ± 0.22 |
6.89 ± 0.992 | 7.789 ± 0.365 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.397 ± 0.167 | 5.932 ± 0.418 |
2.876 ± 0.378 | 4.374 ± 1.049 |
4.374 ± 0.542 | 8.089 ± 0.862 |
5.812 ± 0.806 | 5.153 ± 0.448 |
1.198 ± 0.116 | 6.231 ± 1.397 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
