
Eufriesea mexicana
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Hymenoptera; Apocrita; Aculeata; Apoidea; Apidae; Apinae; Euglossini
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
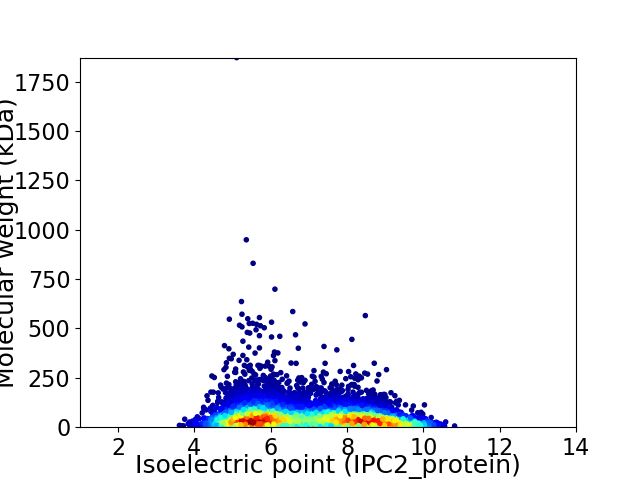
Virtual 2D-PAGE plot for 7068 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A310SKQ8|A0A310SKQ8_9HYME Chloride channel protein OS=Eufriesea mexicana OX=516756 GN=WN48_05835 PE=3 SV=1
MM1 pKa = 8.22DD2 pKa = 3.84MRR4 pKa = 11.84TVTMTRR10 pKa = 11.84GTSTMIIATLTMAMGILTMIIATLTMAMGILTMIIATLTMAMGILTMIIATLTMAMGILTMATGKK75 pKa = 7.42TTMITRR81 pKa = 11.84TSTMVIGTSTIATRR95 pKa = 11.84MSTMATGMSTITIATLTITMRR116 pKa = 11.84TLTMATEE123 pKa = 4.33MSTITIATLTITMTDD138 pKa = 3.49DD139 pKa = 4.16DD140 pKa = 5.1YY141 pKa = 12.21GNIDD145 pKa = 4.82DD146 pKa = 5.93GYY148 pKa = 11.24LDD150 pKa = 3.96VDD152 pKa = 3.59DD153 pKa = 5.18SYY155 pKa = 11.96RR156 pKa = 11.84GFDD159 pKa = 4.61DD160 pKa = 5.9GYY162 pKa = 11.41LDD164 pKa = 4.65VDD166 pKa = 4.23DD167 pKa = 6.2SYY169 pKa = 11.8EE170 pKa = 3.89AWTMATRR177 pKa = 11.84MSTIAVPTSTITMRR191 pKa = 11.84ADD193 pKa = 3.28DD194 pKa = 4.79GYY196 pKa = 11.56RR197 pKa = 11.84DD198 pKa = 3.43VDD200 pKa = 3.49NSYY203 pKa = 11.89GDD205 pKa = 4.24INDD208 pKa = 4.2CYY210 pKa = 11.23EE211 pKa = 4.51DD212 pKa = 3.98VDD214 pKa = 5.54DD215 pKa = 7.04GYY217 pKa = 11.89LDD219 pKa = 3.62VNDD222 pKa = 4.32YY223 pKa = 11.64YY224 pKa = 11.38EE225 pKa = 5.67DD226 pKa = 4.33FDD228 pKa = 5.67DD229 pKa = 5.94GYY231 pKa = 11.69RR232 pKa = 11.84DD233 pKa = 3.42VDD235 pKa = 4.41DD236 pKa = 5.92GYY238 pKa = 11.74LDD240 pKa = 4.89VDD242 pKa = 3.78DD243 pKa = 6.42CYY245 pKa = 11.58GGWTMVTGMATKK257 pKa = 10.15IIATSTITTRR267 pKa = 11.84TLAMATGMSTIATRR281 pKa = 11.84TLTMSTGAGQWLQEE295 pKa = 4.56CRR297 pKa = 11.84RR298 pKa = 11.84CLQGLDD304 pKa = 3.2GSYY307 pKa = 10.97RR308 pKa = 11.84DD309 pKa = 3.68INNSYY314 pKa = 11.1RR315 pKa = 11.84NVNDD319 pKa = 4.27FYY321 pKa = 11.66EE322 pKa = 4.7DD323 pKa = 3.32VDD325 pKa = 4.42DD326 pKa = 4.15GHH328 pKa = 7.82GNVNSVYY335 pKa = 10.4RR336 pKa = 11.84DD337 pKa = 3.41DD338 pKa = 4.26YY339 pKa = 12.01NNYY342 pKa = 10.23EE343 pKa = 4.4YY344 pKa = 11.76VNDD347 pKa = 4.58AYY349 pKa = 11.64SNDD352 pKa = 3.25NVEE355 pKa = 4.83VIGKK359 pKa = 7.51YY360 pKa = 7.76TKK362 pKa = 9.77QWNTDD367 pKa = 2.84
MM1 pKa = 8.22DD2 pKa = 3.84MRR4 pKa = 11.84TVTMTRR10 pKa = 11.84GTSTMIIATLTMAMGILTMIIATLTMAMGILTMIIATLTMAMGILTMIIATLTMAMGILTMATGKK75 pKa = 7.42TTMITRR81 pKa = 11.84TSTMVIGTSTIATRR95 pKa = 11.84MSTMATGMSTITIATLTITMRR116 pKa = 11.84TLTMATEE123 pKa = 4.33MSTITIATLTITMTDD138 pKa = 3.49DD139 pKa = 4.16DD140 pKa = 5.1YY141 pKa = 12.21GNIDD145 pKa = 4.82DD146 pKa = 5.93GYY148 pKa = 11.24LDD150 pKa = 3.96VDD152 pKa = 3.59DD153 pKa = 5.18SYY155 pKa = 11.96RR156 pKa = 11.84GFDD159 pKa = 4.61DD160 pKa = 5.9GYY162 pKa = 11.41LDD164 pKa = 4.65VDD166 pKa = 4.23DD167 pKa = 6.2SYY169 pKa = 11.8EE170 pKa = 3.89AWTMATRR177 pKa = 11.84MSTIAVPTSTITMRR191 pKa = 11.84ADD193 pKa = 3.28DD194 pKa = 4.79GYY196 pKa = 11.56RR197 pKa = 11.84DD198 pKa = 3.43VDD200 pKa = 3.49NSYY203 pKa = 11.89GDD205 pKa = 4.24INDD208 pKa = 4.2CYY210 pKa = 11.23EE211 pKa = 4.51DD212 pKa = 3.98VDD214 pKa = 5.54DD215 pKa = 7.04GYY217 pKa = 11.89LDD219 pKa = 3.62VNDD222 pKa = 4.32YY223 pKa = 11.64YY224 pKa = 11.38EE225 pKa = 5.67DD226 pKa = 4.33FDD228 pKa = 5.67DD229 pKa = 5.94GYY231 pKa = 11.69RR232 pKa = 11.84DD233 pKa = 3.42VDD235 pKa = 4.41DD236 pKa = 5.92GYY238 pKa = 11.74LDD240 pKa = 4.89VDD242 pKa = 3.78DD243 pKa = 6.42CYY245 pKa = 11.58GGWTMVTGMATKK257 pKa = 10.15IIATSTITTRR267 pKa = 11.84TLAMATGMSTIATRR281 pKa = 11.84TLTMSTGAGQWLQEE295 pKa = 4.56CRR297 pKa = 11.84RR298 pKa = 11.84CLQGLDD304 pKa = 3.2GSYY307 pKa = 10.97RR308 pKa = 11.84DD309 pKa = 3.68INNSYY314 pKa = 11.1RR315 pKa = 11.84NVNDD319 pKa = 4.27FYY321 pKa = 11.66EE322 pKa = 4.7DD323 pKa = 3.32VDD325 pKa = 4.42DD326 pKa = 4.15GHH328 pKa = 7.82GNVNSVYY335 pKa = 10.4RR336 pKa = 11.84DD337 pKa = 3.41DD338 pKa = 4.26YY339 pKa = 12.01NNYY342 pKa = 10.23EE343 pKa = 4.4YY344 pKa = 11.76VNDD347 pKa = 4.58AYY349 pKa = 11.64SNDD352 pKa = 3.25NVEE355 pKa = 4.83VIGKK359 pKa = 7.51YY360 pKa = 7.76TKK362 pKa = 9.77QWNTDD367 pKa = 2.84
Molecular weight: 40.62 kDa
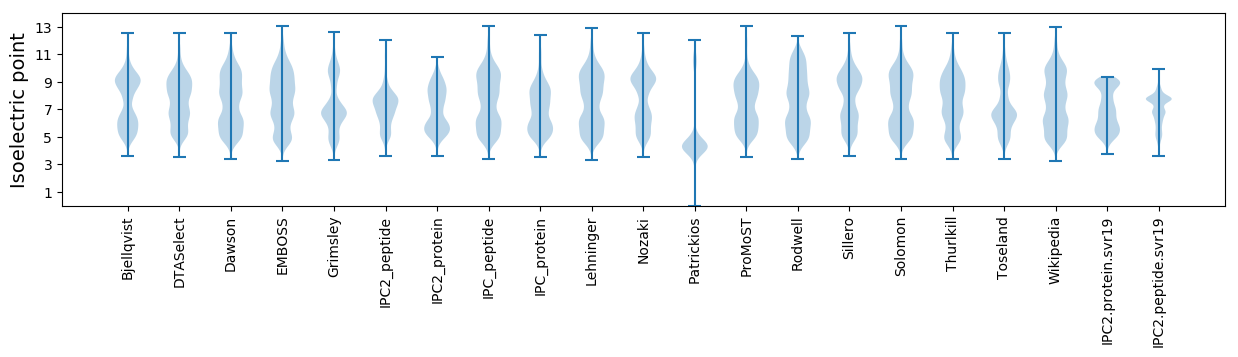
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A310SSF3|A0A310SSF3_9HYME Tubulin alpha chain (Fragment) OS=Eufriesea mexicana OX=516756 GN=WN48_08017 PE=3 SV=1
MM1 pKa = 7.34TGIFKK6 pKa = 9.74TRR8 pKa = 11.84GKK10 pKa = 9.62HH11 pKa = 4.4GKK13 pKa = 8.97RR14 pKa = 11.84ATRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84NVKK23 pKa = 10.06EE24 pKa = 3.48IAGLNNATRR33 pKa = 11.84WKK35 pKa = 10.02RR36 pKa = 11.84KK37 pKa = 5.47QTGGQVPKK45 pKa = 10.31RR46 pKa = 11.84HH47 pKa = 5.9FFHH50 pKa = 7.31GGLAA54 pKa = 3.46
MM1 pKa = 7.34TGIFKK6 pKa = 9.74TRR8 pKa = 11.84GKK10 pKa = 9.62HH11 pKa = 4.4GKK13 pKa = 8.97RR14 pKa = 11.84ATRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84NVKK23 pKa = 10.06EE24 pKa = 3.48IAGLNNATRR33 pKa = 11.84WKK35 pKa = 10.02RR36 pKa = 11.84KK37 pKa = 5.47QTGGQVPKK45 pKa = 10.31RR46 pKa = 11.84HH47 pKa = 5.9FFHH50 pKa = 7.31GGLAA54 pKa = 3.46
Molecular weight: 6.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3581618 |
50 |
16601 |
506.7 |
57.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.903 ± 0.031 | 1.993 ± 0.026 |
5.294 ± 0.024 | 6.983 ± 0.051 |
3.52 ± 0.019 | 5.528 ± 0.035 |
2.468 ± 0.019 | 5.948 ± 0.029 |
6.538 ± 0.044 | 8.946 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.207 ± 0.012 | 5.169 ± 0.032 |
5.036 ± 0.042 | 4.224 ± 0.028 |
5.708 ± 0.031 | 8.202 ± 0.038 |
6.09 ± 0.028 | 6.043 ± 0.028 |
1.102 ± 0.01 | 3.09 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
