
Rose spring dwarf-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
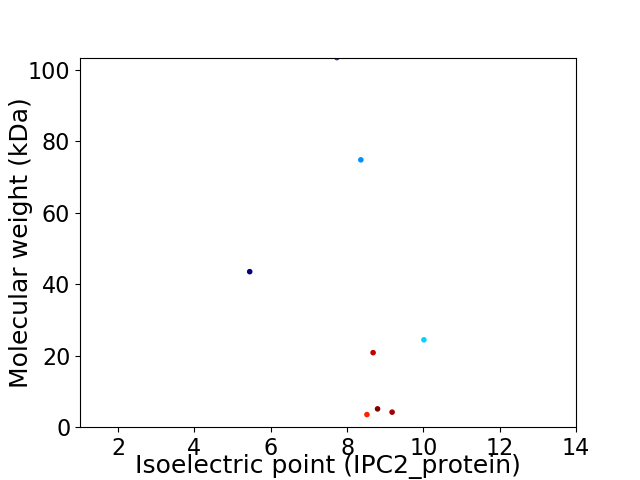
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3FHC0|B3FHC0_9LUTE Readthrough protein OS=Rose spring dwarf-associated virus OX=474454 PE=3 SV=1
MM1 pKa = 7.54LLFEE5 pKa = 5.04GLLTASARR13 pKa = 11.84VVKK16 pKa = 10.59DD17 pKa = 4.59FISYY21 pKa = 9.79IYY23 pKa = 10.61SRR25 pKa = 11.84LKK27 pKa = 10.03SVYY30 pKa = 9.55YY31 pKa = 10.01SLKK34 pKa = 8.92RR35 pKa = 11.84WLWTLQGKK43 pKa = 7.56FQPHH47 pKa = 6.75DD48 pKa = 3.45AFVAMVYY55 pKa = 10.79GYY57 pKa = 9.62MDD59 pKa = 3.98DD60 pKa = 4.75VEE62 pKa = 5.1DD63 pKa = 5.01FEE65 pKa = 5.67VPLALEE71 pKa = 4.25YY72 pKa = 10.71EE73 pKa = 4.37VTEE76 pKa = 4.47RR77 pKa = 11.84EE78 pKa = 4.17LKK80 pKa = 10.01EE81 pKa = 4.2AQDD84 pKa = 3.5EE85 pKa = 4.4LNRR88 pKa = 11.84LEE90 pKa = 4.26SKK92 pKa = 10.76LRR94 pKa = 11.84FTSFNPWVNLVKK106 pKa = 9.99KK107 pKa = 8.85TFTRR111 pKa = 11.84EE112 pKa = 3.67VAPEE116 pKa = 4.09PFWPVPSRR124 pKa = 11.84PDD126 pKa = 3.68CDD128 pKa = 4.24DD129 pKa = 3.91PCISSPLIEE138 pKa = 5.12DD139 pKa = 3.8EE140 pKa = 4.77EE141 pKa = 4.62EE142 pKa = 4.34VSGPEE147 pKa = 3.87PEE149 pKa = 5.71PIFSQQKK156 pKa = 9.65LDD158 pKa = 3.55EE159 pKa = 4.55RR160 pKa = 11.84LCMEE164 pKa = 4.56ARR166 pKa = 11.84MDD168 pKa = 3.75VLNSEE173 pKa = 4.41VKK175 pKa = 9.66FDD177 pKa = 4.01KK178 pKa = 10.74VRR180 pKa = 11.84EE181 pKa = 4.22RR182 pKa = 11.84INIVYY187 pKa = 10.2QEE189 pKa = 4.05VRR191 pKa = 11.84GARR194 pKa = 11.84PFGKK198 pKa = 9.76IFNTMHH204 pKa = 6.11QRR206 pKa = 11.84MKK208 pKa = 10.83HH209 pKa = 4.65VGKK212 pKa = 10.05CLKK215 pKa = 10.16RR216 pKa = 11.84RR217 pKa = 11.84SEE219 pKa = 3.75ASARR223 pKa = 11.84SVEE226 pKa = 4.02FEE228 pKa = 3.88KK229 pKa = 10.81RR230 pKa = 11.84VNAKK234 pKa = 9.05TDD236 pKa = 2.96IADD239 pKa = 3.67FHH241 pKa = 6.74SLCEE245 pKa = 4.11VEE247 pKa = 4.82EE248 pKa = 4.55VEE250 pKa = 4.29TGEE253 pKa = 3.99FHH255 pKa = 7.53PVKK258 pKa = 10.34QDD260 pKa = 3.09ADD262 pKa = 4.07GEE264 pKa = 4.33DD265 pKa = 4.08LPRR268 pKa = 11.84LPKK271 pKa = 9.97IEE273 pKa = 3.98VVRR276 pKa = 11.84RR277 pKa = 11.84IKK279 pKa = 10.71DD280 pKa = 3.32NCHH283 pKa = 6.05RR284 pKa = 11.84PAATWIRR291 pKa = 11.84EE292 pKa = 3.86YY293 pKa = 11.12VRR295 pKa = 11.84CKK297 pKa = 10.57NSQLSADD304 pKa = 4.17EE305 pKa = 4.43VSHH308 pKa = 5.81ATITRR313 pKa = 11.84YY314 pKa = 9.75VEE316 pKa = 3.98QFCEE320 pKa = 4.1KK321 pKa = 10.81NKK323 pKa = 9.81MDD325 pKa = 3.45MDD327 pKa = 3.85SRR329 pKa = 11.84VFLMQRR335 pKa = 11.84ALLMVPIPKK344 pKa = 9.98PIDD347 pKa = 3.01IDD349 pKa = 3.15IAMTVHH355 pKa = 6.45SPAARR360 pKa = 11.84EE361 pKa = 3.78LRR363 pKa = 11.84SIVDD367 pKa = 3.74TAASSVFF374 pKa = 3.36
MM1 pKa = 7.54LLFEE5 pKa = 5.04GLLTASARR13 pKa = 11.84VVKK16 pKa = 10.59DD17 pKa = 4.59FISYY21 pKa = 9.79IYY23 pKa = 10.61SRR25 pKa = 11.84LKK27 pKa = 10.03SVYY30 pKa = 9.55YY31 pKa = 10.01SLKK34 pKa = 8.92RR35 pKa = 11.84WLWTLQGKK43 pKa = 7.56FQPHH47 pKa = 6.75DD48 pKa = 3.45AFVAMVYY55 pKa = 10.79GYY57 pKa = 9.62MDD59 pKa = 3.98DD60 pKa = 4.75VEE62 pKa = 5.1DD63 pKa = 5.01FEE65 pKa = 5.67VPLALEE71 pKa = 4.25YY72 pKa = 10.71EE73 pKa = 4.37VTEE76 pKa = 4.47RR77 pKa = 11.84EE78 pKa = 4.17LKK80 pKa = 10.01EE81 pKa = 4.2AQDD84 pKa = 3.5EE85 pKa = 4.4LNRR88 pKa = 11.84LEE90 pKa = 4.26SKK92 pKa = 10.76LRR94 pKa = 11.84FTSFNPWVNLVKK106 pKa = 9.99KK107 pKa = 8.85TFTRR111 pKa = 11.84EE112 pKa = 3.67VAPEE116 pKa = 4.09PFWPVPSRR124 pKa = 11.84PDD126 pKa = 3.68CDD128 pKa = 4.24DD129 pKa = 3.91PCISSPLIEE138 pKa = 5.12DD139 pKa = 3.8EE140 pKa = 4.77EE141 pKa = 4.62EE142 pKa = 4.34VSGPEE147 pKa = 3.87PEE149 pKa = 5.71PIFSQQKK156 pKa = 9.65LDD158 pKa = 3.55EE159 pKa = 4.55RR160 pKa = 11.84LCMEE164 pKa = 4.56ARR166 pKa = 11.84MDD168 pKa = 3.75VLNSEE173 pKa = 4.41VKK175 pKa = 9.66FDD177 pKa = 4.01KK178 pKa = 10.74VRR180 pKa = 11.84EE181 pKa = 4.22RR182 pKa = 11.84INIVYY187 pKa = 10.2QEE189 pKa = 4.05VRR191 pKa = 11.84GARR194 pKa = 11.84PFGKK198 pKa = 9.76IFNTMHH204 pKa = 6.11QRR206 pKa = 11.84MKK208 pKa = 10.83HH209 pKa = 4.65VGKK212 pKa = 10.05CLKK215 pKa = 10.16RR216 pKa = 11.84RR217 pKa = 11.84SEE219 pKa = 3.75ASARR223 pKa = 11.84SVEE226 pKa = 4.02FEE228 pKa = 3.88KK229 pKa = 10.81RR230 pKa = 11.84VNAKK234 pKa = 9.05TDD236 pKa = 2.96IADD239 pKa = 3.67FHH241 pKa = 6.74SLCEE245 pKa = 4.11VEE247 pKa = 4.82EE248 pKa = 4.55VEE250 pKa = 4.29TGEE253 pKa = 3.99FHH255 pKa = 7.53PVKK258 pKa = 10.34QDD260 pKa = 3.09ADD262 pKa = 4.07GEE264 pKa = 4.33DD265 pKa = 4.08LPRR268 pKa = 11.84LPKK271 pKa = 9.97IEE273 pKa = 3.98VVRR276 pKa = 11.84RR277 pKa = 11.84IKK279 pKa = 10.71DD280 pKa = 3.32NCHH283 pKa = 6.05RR284 pKa = 11.84PAATWIRR291 pKa = 11.84EE292 pKa = 3.86YY293 pKa = 11.12VRR295 pKa = 11.84CKK297 pKa = 10.57NSQLSADD304 pKa = 4.17EE305 pKa = 4.43VSHH308 pKa = 5.81ATITRR313 pKa = 11.84YY314 pKa = 9.75VEE316 pKa = 3.98QFCEE320 pKa = 4.1KK321 pKa = 10.81NKK323 pKa = 9.81MDD325 pKa = 3.45MDD327 pKa = 3.85SRR329 pKa = 11.84VFLMQRR335 pKa = 11.84ALLMVPIPKK344 pKa = 9.98PIDD347 pKa = 3.01IDD349 pKa = 3.15IAMTVHH355 pKa = 6.45SPAARR360 pKa = 11.84EE361 pKa = 3.78LRR363 pKa = 11.84SIVDD367 pKa = 3.74TAASSVFF374 pKa = 3.36
Molecular weight: 43.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3FHC2|B3FHC2_9LUTE Movement protein OS=Rose spring dwarf-associated virus OX=474454 PE=3 SV=1
MM1 pKa = 6.6STVVVRR7 pKa = 11.84QQARR11 pKa = 11.84NNSRR15 pKa = 11.84RR16 pKa = 11.84NGQAQQAQGRR26 pKa = 11.84SRR28 pKa = 11.84QPNKK32 pKa = 10.02ARR34 pKa = 11.84PVVVQVQPSRR44 pKa = 11.84NGRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GGRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84GSRR64 pKa = 11.84MASSRR69 pKa = 11.84SHH71 pKa = 5.36WEE73 pKa = 3.84DD74 pKa = 3.21YY75 pKa = 11.21KK76 pKa = 10.93FTINNLKK83 pKa = 10.64ASDD86 pKa = 3.55AGVVKK91 pKa = 9.97FGPSISQCSALKK103 pKa = 10.69SGIFKK108 pKa = 10.61SFHH111 pKa = 5.24EE112 pKa = 4.58FKK114 pKa = 9.91ITNLNVKK121 pKa = 9.61YY122 pKa = 8.8ITHH125 pKa = 6.99AASTTSGAFAVEE137 pKa = 4.15VDD139 pKa = 4.14TSCTQTTLKK148 pKa = 10.63SYY150 pKa = 10.44LQTVPVAKK158 pKa = 9.99CGQFSWPAGKK168 pKa = 9.7IRR170 pKa = 11.84GTGWLPTPDD179 pKa = 4.0PDD181 pKa = 4.09KK182 pKa = 11.37TPVDD186 pKa = 3.63KK187 pKa = 11.29DD188 pKa = 3.39NQFFLLYY195 pKa = 10.25AGNGPSSVAGQFVITARR212 pKa = 11.84CWFQSPRR219 pKa = 11.84EE220 pKa = 4.11
MM1 pKa = 6.6STVVVRR7 pKa = 11.84QQARR11 pKa = 11.84NNSRR15 pKa = 11.84RR16 pKa = 11.84NGQAQQAQGRR26 pKa = 11.84SRR28 pKa = 11.84QPNKK32 pKa = 10.02ARR34 pKa = 11.84PVVVQVQPSRR44 pKa = 11.84NGRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84GGRR55 pKa = 11.84RR56 pKa = 11.84SSRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84GSRR64 pKa = 11.84MASSRR69 pKa = 11.84SHH71 pKa = 5.36WEE73 pKa = 3.84DD74 pKa = 3.21YY75 pKa = 11.21KK76 pKa = 10.93FTINNLKK83 pKa = 10.64ASDD86 pKa = 3.55AGVVKK91 pKa = 9.97FGPSISQCSALKK103 pKa = 10.69SGIFKK108 pKa = 10.61SFHH111 pKa = 5.24EE112 pKa = 4.58FKK114 pKa = 9.91ITNLNVKK121 pKa = 9.61YY122 pKa = 8.8ITHH125 pKa = 6.99AASTTSGAFAVEE137 pKa = 4.15VDD139 pKa = 4.14TSCTQTTLKK148 pKa = 10.63SYY150 pKa = 10.44LQTVPVAKK158 pKa = 9.99CGQFSWPAGKK168 pKa = 9.7IRR170 pKa = 11.84GTGWLPTPDD179 pKa = 4.0PDD181 pKa = 4.09KK182 pKa = 11.37TPVDD186 pKa = 3.63KK187 pKa = 11.29DD188 pKa = 3.39NQFFLLYY195 pKa = 10.25AGNGPSSVAGQFVITARR212 pKa = 11.84CWFQSPRR219 pKa = 11.84EE220 pKa = 4.11
Molecular weight: 24.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2474 |
32 |
901 |
309.3 |
34.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.346 ± 0.501 | 1.778 ± 0.239 |
5.457 ± 0.419 | 6.144 ± 1.071 |
4.568 ± 0.388 | 5.295 ± 0.757 |
2.142 ± 0.206 | 4.406 ± 0.529 |
5.659 ± 0.511 | 7.599 ± 0.803 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.344 ± 0.56 | 3.88 ± 0.412 |
6.669 ± 0.751 | 4.244 ± 0.609 |
7.922 ± 0.326 | 8.488 ± 0.876 |
5.053 ± 0.828 | 8.084 ± 0.319 |
1.334 ± 0.172 | 2.546 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
