
Salmo trutta (Brown trout)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Protacanthopterygii; Salmoniformes; Salmonidae;
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
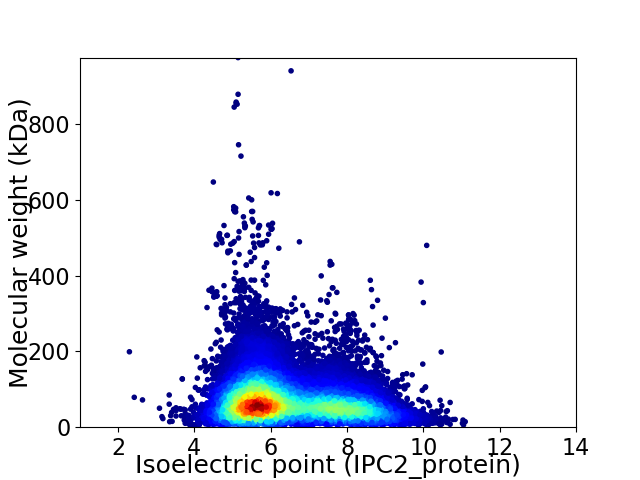
Virtual 2D-PAGE plot for 114752 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A673WCC1|A0A673WCC1_SALTR Uncharacterized protein OS=Salmo trutta OX=8032 PE=4 SV=1
MM1 pKa = 7.69KK2 pKa = 10.17IIILLIIMVVSMLILLVIIIMIAVVMVVSMLVMIIMIEE40 pKa = 4.08VVMVVSMLMLIMIAVVMEE58 pKa = 3.94VSADD62 pKa = 3.38VDD64 pKa = 4.36DD65 pKa = 5.1NDD67 pKa = 4.89SGGNGSKK74 pKa = 10.87YY75 pKa = 10.9ADD77 pKa = 4.19DD78 pKa = 5.75DD79 pKa = 4.79YY80 pKa = 11.85NDD82 pKa = 3.59SGGNGSKK89 pKa = 10.84YY90 pKa = 10.75VDD92 pKa = 5.3DD93 pKa = 6.08DD94 pKa = 5.0DD95 pKa = 6.6DD96 pKa = 5.5YY97 pKa = 12.06NDD99 pKa = 3.64SGGNGSKK106 pKa = 10.79YY107 pKa = 10.94ADD109 pKa = 3.79VDD111 pKa = 3.75DD112 pKa = 6.31DD113 pKa = 4.89GYY115 pKa = 11.66NDD117 pKa = 3.58SGGNGSKK124 pKa = 10.79YY125 pKa = 10.95ADD127 pKa = 3.37VDD129 pKa = 3.49DD130 pKa = 5.6DD131 pKa = 5.13YY132 pKa = 12.08NVSKK136 pKa = 11.04YY137 pKa = 11.24ADD139 pKa = 3.73VDD141 pKa = 3.81DD142 pKa = 6.45DD143 pKa = 5.25DD144 pKa = 6.26YY145 pKa = 12.03NDD147 pKa = 3.58SGGNGSKK154 pKa = 10.79YY155 pKa = 10.97ADD157 pKa = 4.02VDD159 pKa = 3.83DD160 pKa = 6.45DD161 pKa = 5.25DD162 pKa = 6.26YY163 pKa = 12.03NDD165 pKa = 3.58SGGNGSKK172 pKa = 10.79YY173 pKa = 10.95ADD175 pKa = 3.54VDD177 pKa = 3.82DD178 pKa = 5.93DD179 pKa = 4.88YY180 pKa = 11.94NDD182 pKa = 3.5SGGNGSKK189 pKa = 10.86YY190 pKa = 10.82VDD192 pKa = 4.18DD193 pKa = 6.01DD194 pKa = 4.79YY195 pKa = 11.99NDD197 pKa = 3.51SGGNGSKK204 pKa = 10.79YY205 pKa = 10.95ADD207 pKa = 3.54VDD209 pKa = 3.82DD210 pKa = 5.93DD211 pKa = 4.88YY212 pKa = 11.94NDD214 pKa = 3.5SGGNGSKK221 pKa = 10.79YY222 pKa = 10.97ADD224 pKa = 3.05VDD226 pKa = 4.14DD227 pKa = 5.19YY228 pKa = 11.92NDD230 pKa = 3.47SGGNGSKK237 pKa = 10.79YY238 pKa = 10.9ADD240 pKa = 3.29VDD242 pKa = 4.22DD243 pKa = 5.19YY244 pKa = 11.86SDD246 pKa = 3.57SGGNGSKK253 pKa = 10.79YY254 pKa = 10.95ADD256 pKa = 3.54VDD258 pKa = 3.82DD259 pKa = 5.93DD260 pKa = 4.88YY261 pKa = 11.94NDD263 pKa = 3.5SGGNGSKK270 pKa = 10.84YY271 pKa = 10.77VDD273 pKa = 5.43DD274 pKa = 6.82DD275 pKa = 4.95DD276 pKa = 7.34DD277 pKa = 5.69DD278 pKa = 5.8YY279 pKa = 12.09NDD281 pKa = 3.64SGGNGSKK288 pKa = 10.87YY289 pKa = 10.9ADD291 pKa = 4.19DD292 pKa = 5.75DD293 pKa = 4.79YY294 pKa = 11.85NDD296 pKa = 3.59SGGNGSKK303 pKa = 10.48YY304 pKa = 10.76AAVDD308 pKa = 3.78YY309 pKa = 11.05NDD311 pKa = 3.61SGGNGSKK318 pKa = 10.79YY319 pKa = 10.9ADD321 pKa = 3.29VDD323 pKa = 4.22DD324 pKa = 5.19YY325 pKa = 11.86SDD327 pKa = 3.57SGGNGSKK334 pKa = 10.79YY335 pKa = 10.95ADD337 pKa = 3.54VDD339 pKa = 3.82DD340 pKa = 5.93DD341 pKa = 4.88YY342 pKa = 11.94NDD344 pKa = 3.5SGGNGSKK351 pKa = 10.53YY352 pKa = 10.61AADD355 pKa = 3.3VDD357 pKa = 4.74DD358 pKa = 5.37YY359 pKa = 11.77NDD361 pKa = 3.42SSGNGSKK368 pKa = 10.58YY369 pKa = 10.64AADD372 pKa = 3.3VDD374 pKa = 4.69DD375 pKa = 5.21YY376 pKa = 11.73NDD378 pKa = 3.47SGGNGSKK385 pKa = 10.53YY386 pKa = 10.61AADD389 pKa = 3.3VDD391 pKa = 4.69DD392 pKa = 5.21YY393 pKa = 11.73NDD395 pKa = 3.47SGGNGSKK402 pKa = 10.53YY403 pKa = 10.61AADD406 pKa = 3.3VDD408 pKa = 4.69DD409 pKa = 5.21YY410 pKa = 11.73NDD412 pKa = 3.47SGGNGSKK419 pKa = 10.78YY420 pKa = 10.8ADD422 pKa = 3.53GDD424 pKa = 3.83GSQTVMAHH432 pKa = 6.25GCHH435 pKa = 6.12LTDD438 pKa = 4.62EE439 pKa = 4.32EE440 pKa = 4.59LKK442 pKa = 10.94LFRR445 pKa = 11.84EE446 pKa = 4.42KK447 pKa = 10.72GASVAHH453 pKa = 6.45CPNSNISLCSGMLDD467 pKa = 3.1ARR469 pKa = 11.84RR470 pKa = 11.84VLNHH474 pKa = 6.11KK475 pKa = 10.12VKK477 pKa = 10.81LGLGTDD483 pKa = 3.48VAGGYY488 pKa = 9.17SPSMLDD494 pKa = 3.18AVRR497 pKa = 11.84RR498 pKa = 11.84TLDD501 pKa = 3.04TSKK504 pKa = 11.19ALTIQDD510 pKa = 3.78PQYY513 pKa = 9.75QTLTFEE519 pKa = 4.37EE520 pKa = 4.89VFRR523 pKa = 11.84LATLGGSQALSLDD536 pKa = 3.35EE537 pKa = 4.16HH538 pKa = 6.42TGNFQVGKK546 pKa = 10.13DD547 pKa = 3.55FDD549 pKa = 4.05ALRR552 pKa = 11.84VNAAMEE558 pKa = 4.55GGPIDD563 pKa = 5.76LINHH567 pKa = 5.97GEE569 pKa = 4.23GPKK572 pKa = 10.38VLLEE576 pKa = 4.17KK577 pKa = 10.48FLNLGDD583 pKa = 4.19DD584 pKa = 3.68RR585 pKa = 11.84NIAEE589 pKa = 4.53VYY591 pKa = 9.15VAGKK595 pKa = 9.83RR596 pKa = 11.84VVPFVEE602 pKa = 4.69RR603 pKa = 11.84PP604 pKa = 3.17
MM1 pKa = 7.69KK2 pKa = 10.17IIILLIIMVVSMLILLVIIIMIAVVMVVSMLVMIIMIEE40 pKa = 4.08VVMVVSMLMLIMIAVVMEE58 pKa = 3.94VSADD62 pKa = 3.38VDD64 pKa = 4.36DD65 pKa = 5.1NDD67 pKa = 4.89SGGNGSKK74 pKa = 10.87YY75 pKa = 10.9ADD77 pKa = 4.19DD78 pKa = 5.75DD79 pKa = 4.79YY80 pKa = 11.85NDD82 pKa = 3.59SGGNGSKK89 pKa = 10.84YY90 pKa = 10.75VDD92 pKa = 5.3DD93 pKa = 6.08DD94 pKa = 5.0DD95 pKa = 6.6DD96 pKa = 5.5YY97 pKa = 12.06NDD99 pKa = 3.64SGGNGSKK106 pKa = 10.79YY107 pKa = 10.94ADD109 pKa = 3.79VDD111 pKa = 3.75DD112 pKa = 6.31DD113 pKa = 4.89GYY115 pKa = 11.66NDD117 pKa = 3.58SGGNGSKK124 pKa = 10.79YY125 pKa = 10.95ADD127 pKa = 3.37VDD129 pKa = 3.49DD130 pKa = 5.6DD131 pKa = 5.13YY132 pKa = 12.08NVSKK136 pKa = 11.04YY137 pKa = 11.24ADD139 pKa = 3.73VDD141 pKa = 3.81DD142 pKa = 6.45DD143 pKa = 5.25DD144 pKa = 6.26YY145 pKa = 12.03NDD147 pKa = 3.58SGGNGSKK154 pKa = 10.79YY155 pKa = 10.97ADD157 pKa = 4.02VDD159 pKa = 3.83DD160 pKa = 6.45DD161 pKa = 5.25DD162 pKa = 6.26YY163 pKa = 12.03NDD165 pKa = 3.58SGGNGSKK172 pKa = 10.79YY173 pKa = 10.95ADD175 pKa = 3.54VDD177 pKa = 3.82DD178 pKa = 5.93DD179 pKa = 4.88YY180 pKa = 11.94NDD182 pKa = 3.5SGGNGSKK189 pKa = 10.86YY190 pKa = 10.82VDD192 pKa = 4.18DD193 pKa = 6.01DD194 pKa = 4.79YY195 pKa = 11.99NDD197 pKa = 3.51SGGNGSKK204 pKa = 10.79YY205 pKa = 10.95ADD207 pKa = 3.54VDD209 pKa = 3.82DD210 pKa = 5.93DD211 pKa = 4.88YY212 pKa = 11.94NDD214 pKa = 3.5SGGNGSKK221 pKa = 10.79YY222 pKa = 10.97ADD224 pKa = 3.05VDD226 pKa = 4.14DD227 pKa = 5.19YY228 pKa = 11.92NDD230 pKa = 3.47SGGNGSKK237 pKa = 10.79YY238 pKa = 10.9ADD240 pKa = 3.29VDD242 pKa = 4.22DD243 pKa = 5.19YY244 pKa = 11.86SDD246 pKa = 3.57SGGNGSKK253 pKa = 10.79YY254 pKa = 10.95ADD256 pKa = 3.54VDD258 pKa = 3.82DD259 pKa = 5.93DD260 pKa = 4.88YY261 pKa = 11.94NDD263 pKa = 3.5SGGNGSKK270 pKa = 10.84YY271 pKa = 10.77VDD273 pKa = 5.43DD274 pKa = 6.82DD275 pKa = 4.95DD276 pKa = 7.34DD277 pKa = 5.69DD278 pKa = 5.8YY279 pKa = 12.09NDD281 pKa = 3.64SGGNGSKK288 pKa = 10.87YY289 pKa = 10.9ADD291 pKa = 4.19DD292 pKa = 5.75DD293 pKa = 4.79YY294 pKa = 11.85NDD296 pKa = 3.59SGGNGSKK303 pKa = 10.48YY304 pKa = 10.76AAVDD308 pKa = 3.78YY309 pKa = 11.05NDD311 pKa = 3.61SGGNGSKK318 pKa = 10.79YY319 pKa = 10.9ADD321 pKa = 3.29VDD323 pKa = 4.22DD324 pKa = 5.19YY325 pKa = 11.86SDD327 pKa = 3.57SGGNGSKK334 pKa = 10.79YY335 pKa = 10.95ADD337 pKa = 3.54VDD339 pKa = 3.82DD340 pKa = 5.93DD341 pKa = 4.88YY342 pKa = 11.94NDD344 pKa = 3.5SGGNGSKK351 pKa = 10.53YY352 pKa = 10.61AADD355 pKa = 3.3VDD357 pKa = 4.74DD358 pKa = 5.37YY359 pKa = 11.77NDD361 pKa = 3.42SSGNGSKK368 pKa = 10.58YY369 pKa = 10.64AADD372 pKa = 3.3VDD374 pKa = 4.69DD375 pKa = 5.21YY376 pKa = 11.73NDD378 pKa = 3.47SGGNGSKK385 pKa = 10.53YY386 pKa = 10.61AADD389 pKa = 3.3VDD391 pKa = 4.69DD392 pKa = 5.21YY393 pKa = 11.73NDD395 pKa = 3.47SGGNGSKK402 pKa = 10.53YY403 pKa = 10.61AADD406 pKa = 3.3VDD408 pKa = 4.69DD409 pKa = 5.21YY410 pKa = 11.73NDD412 pKa = 3.47SGGNGSKK419 pKa = 10.78YY420 pKa = 10.8ADD422 pKa = 3.53GDD424 pKa = 3.83GSQTVMAHH432 pKa = 6.25GCHH435 pKa = 6.12LTDD438 pKa = 4.62EE439 pKa = 4.32EE440 pKa = 4.59LKK442 pKa = 10.94LFRR445 pKa = 11.84EE446 pKa = 4.42KK447 pKa = 10.72GASVAHH453 pKa = 6.45CPNSNISLCSGMLDD467 pKa = 3.1ARR469 pKa = 11.84RR470 pKa = 11.84VLNHH474 pKa = 6.11KK475 pKa = 10.12VKK477 pKa = 10.81LGLGTDD483 pKa = 3.48VAGGYY488 pKa = 9.17SPSMLDD494 pKa = 3.18AVRR497 pKa = 11.84RR498 pKa = 11.84TLDD501 pKa = 3.04TSKK504 pKa = 11.19ALTIQDD510 pKa = 3.78PQYY513 pKa = 9.75QTLTFEE519 pKa = 4.37EE520 pKa = 4.89VFRR523 pKa = 11.84LATLGGSQALSLDD536 pKa = 3.35EE537 pKa = 4.16HH538 pKa = 6.42TGNFQVGKK546 pKa = 10.13DD547 pKa = 3.55FDD549 pKa = 4.05ALRR552 pKa = 11.84VNAAMEE558 pKa = 4.55GGPIDD563 pKa = 5.76LINHH567 pKa = 5.97GEE569 pKa = 4.23GPKK572 pKa = 10.38VLLEE576 pKa = 4.17KK577 pKa = 10.48FLNLGDD583 pKa = 4.19DD584 pKa = 3.68RR585 pKa = 11.84NIAEE589 pKa = 4.53VYY591 pKa = 9.15VAGKK595 pKa = 9.83RR596 pKa = 11.84VVPFVEE602 pKa = 4.69RR603 pKa = 11.84PP604 pKa = 3.17
Molecular weight: 63.96 kDa
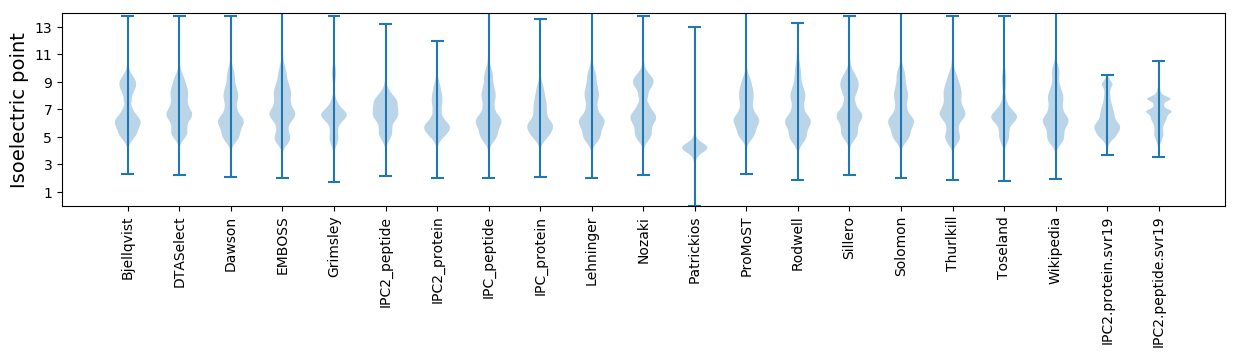
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A674AW04|A0A674AW04_SALTR Leiomodin-2-like OS=Salmo trutta OX=8032 GN=LOC115198757 PE=4 SV=1
KKK2 pKa = 10.25VLCGPARR9 pKa = 11.84SSGSRR14 pKa = 11.84RR15 pKa = 11.84GPLALGAVLWLSARR29 pKa = 11.84PVVLRR34 pKa = 11.84PVVLRR39 pKa = 11.84PVVLRR44 pKa = 11.84PVVLRR49 pKa = 11.84PVVLRR54 pKa = 11.84PVVLRR59 pKa = 11.84PVVLRR64 pKa = 11.84PVVLRR69 pKa = 11.84PVVLRR74 pKa = 11.84PVVLRR79 pKa = 11.84PVVLRR84 pKa = 11.84PVVLRR89 pKa = 11.84PVVLRR94 pKa = 11.84PVVLRR99 pKa = 11.84PVVLRR104 pKa = 11.84PVVLRR109 pKa = 11.84PVVLRR114 pKa = 11.84PVVLRR119 pKa = 11.84PVVLRR124 pKa = 11.84PVVLRR129 pKa = 11.84PVVLRR134 pKa = 11.84PVVLM
KKK2 pKa = 10.25VLCGPARR9 pKa = 11.84SSGSRR14 pKa = 11.84RR15 pKa = 11.84GPLALGAVLWLSARR29 pKa = 11.84PVVLRR34 pKa = 11.84PVVLRR39 pKa = 11.84PVVLRR44 pKa = 11.84PVVLRR49 pKa = 11.84PVVLRR54 pKa = 11.84PVVLRR59 pKa = 11.84PVVLRR64 pKa = 11.84PVVLRR69 pKa = 11.84PVVLRR74 pKa = 11.84PVVLRR79 pKa = 11.84PVVLRR84 pKa = 11.84PVVLRR89 pKa = 11.84PVVLRR94 pKa = 11.84PVVLRR99 pKa = 11.84PVVLRR104 pKa = 11.84PVVLRR109 pKa = 11.84PVVLRR114 pKa = 11.84PVVLRR119 pKa = 11.84PVVLRR124 pKa = 11.84PVVLRR129 pKa = 11.84PVVLRR134 pKa = 11.84PVVLM
Molecular weight: 15.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
79092270 |
18 |
8935 |
689.2 |
77.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.245 ± 0.006 | 2.327 ± 0.007 |
5.201 ± 0.004 | 6.787 ± 0.01 |
3.637 ± 0.005 | 6.531 ± 0.008 |
2.747 ± 0.004 | 4.465 ± 0.005 |
5.525 ± 0.008 | 9.645 ± 0.009 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.003 | 3.872 ± 0.004 |
5.643 ± 0.009 | 4.703 ± 0.006 |
5.558 ± 0.006 | 8.306 ± 0.008 |
5.817 ± 0.008 | 6.474 ± 0.005 |
1.167 ± 0.002 | 2.875 ± 0.004 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
