
Jannaschia helgolandensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Jannaschia
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
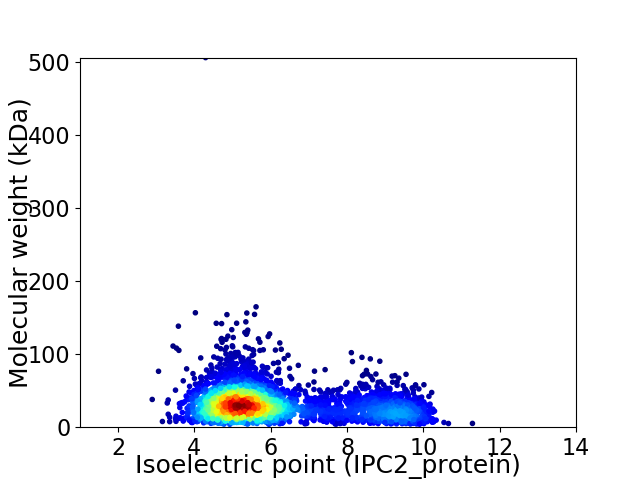
Virtual 2D-PAGE plot for 3471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7G164|A0A1H7G164_9RHOB Farnesyl-diphosphate synthase OS=Jannaschia helgolandensis OX=188906 GN=SAMN04488526_0275 PE=3 SV=1
MM1 pKa = 7.15PTIDD5 pKa = 4.75GSISDD10 pKa = 4.04WSPDD14 pKa = 3.14ARR16 pKa = 11.84LDD18 pKa = 3.6TAATGTLGYY27 pKa = 9.98GLWGVADD34 pKa = 3.74PDD36 pKa = 3.72YY37 pKa = 10.8FYY39 pKa = 11.2FAITSDD45 pKa = 3.56DD46 pKa = 4.01VIIGEE51 pKa = 4.38NTTLWLDD58 pKa = 3.38TDD60 pKa = 4.52LDD62 pKa = 4.15RR63 pKa = 11.84GTGYY67 pKa = 10.33QIWGWAGGAEE77 pKa = 4.23YY78 pKa = 10.1QVEE81 pKa = 4.08IAADD85 pKa = 3.77GTARR89 pKa = 11.84LYY91 pKa = 11.13SGGPEE96 pKa = 3.79EE97 pKa = 4.18TLVAEE102 pKa = 4.59LEE104 pKa = 4.44AVRR107 pKa = 11.84SADD110 pKa = 3.14GRR112 pKa = 11.84VLEE115 pKa = 4.11FRR117 pKa = 11.84IDD119 pKa = 3.51RR120 pKa = 11.84SLLAGNPDD128 pKa = 3.47AVRR131 pKa = 11.84VYY133 pKa = 11.62ADD135 pKa = 3.52VNDD138 pKa = 4.12SAFLPNSYY146 pKa = 11.09ANTNFIVAEE155 pKa = 4.11PSPSVVTGGKK165 pKa = 7.65TLDD168 pKa = 3.85GNLVDD173 pKa = 3.99WDD175 pKa = 3.6AATRR179 pKa = 11.84LDD181 pKa = 3.95TPATGAAGYY190 pKa = 10.63AFYY193 pKa = 11.34GDD195 pKa = 3.83LQDD198 pKa = 4.49NVFTFAISTDD208 pKa = 3.18GTAIGANTTIWLDD221 pKa = 3.44TDD223 pKa = 3.9RR224 pKa = 11.84DD225 pKa = 3.89TATGYY230 pKa = 8.53QVWGWAVGGEE240 pKa = 4.05YY241 pKa = 10.42NINITADD248 pKa = 3.38GEE250 pKa = 4.19ARR252 pKa = 11.84LYY254 pKa = 10.24TGAAGEE260 pKa = 4.38TFVANLDD267 pKa = 3.43FAYY270 pKa = 10.71NADD273 pKa = 3.52GTVIEE278 pKa = 4.49IALDD282 pKa = 3.45RR283 pKa = 11.84ALVGNPASMRR293 pKa = 11.84VFADD297 pKa = 3.29VNDD300 pKa = 3.96SVFIPNSYY308 pKa = 11.22ANANFIVGSEE318 pKa = 4.01PTPVIGGITLDD329 pKa = 3.78GSVSEE334 pKa = 4.39WTPEE338 pKa = 4.57SILLTQGGYY347 pKa = 6.9QLRR350 pKa = 11.84GEE352 pKa = 4.53FVVDD356 pKa = 3.65AAGDD360 pKa = 3.58QYY362 pKa = 11.94VFAIEE367 pKa = 4.51SDD369 pKa = 3.43GTTIGANTTIWLDD382 pKa = 3.14TDD384 pKa = 4.36LNSTTGYY391 pKa = 10.2QIWGWAGGAEE401 pKa = 4.19YY402 pKa = 10.63NVNITADD409 pKa = 3.62GQARR413 pKa = 11.84LYY415 pKa = 10.83SGAAGGTYY423 pKa = 9.95IADD426 pKa = 3.77LDD428 pKa = 4.35FGFSADD434 pKa = 3.38GTGFEE439 pKa = 4.36VAISKK444 pKa = 10.45DD445 pKa = 4.02LLSGAPDD452 pKa = 3.35QVRR455 pKa = 11.84ILADD459 pKa = 3.3VNDD462 pKa = 4.1AVFLPGDD469 pKa = 3.63YY470 pKa = 10.8SSANLVVGAPLPPPTDD486 pKa = 3.52APEE489 pKa = 3.74SRR491 pKa = 11.84IAIVYY496 pKa = 9.67SATTAANFYY505 pKa = 10.71DD506 pKa = 3.16ITAYY510 pKa = 10.4GQLFMAAQNQAMQAGIPFDD529 pKa = 4.5LLTEE533 pKa = 4.69ADD535 pKa = 4.33LLDD538 pKa = 3.9PTGLAGYY545 pKa = 9.43DD546 pKa = 3.29AIVFPGFSNVQASQAATIAASLTTAAQDD574 pKa = 3.52YY575 pKa = 8.71GVGMIVAGNFLTNDD589 pKa = 3.34EE590 pKa = 4.66TGAALPGNSYY600 pKa = 11.51ARR602 pKa = 11.84MNALLGVTLEE612 pKa = 4.69GFGATQGITLNASNAANPILDD633 pKa = 4.61GYY635 pKa = 9.62TNGQVVGNYY644 pKa = 10.39GNLSYY649 pKa = 11.58LNFTDD654 pKa = 3.72TTGSGQVLFEE664 pKa = 4.12QVVSDD669 pKa = 3.62GTYY672 pKa = 10.28DD673 pKa = 4.07AVIATQTAGRR683 pKa = 11.84NVHH686 pKa = 6.51FATDD690 pKa = 4.23AIMGNNNILGEE701 pKa = 4.47AIDD704 pKa = 5.3WVLQDD709 pKa = 3.75NAPDD713 pKa = 3.31ISLLMTRR720 pKa = 11.84NSSIFYY726 pKa = 10.21SRR728 pKa = 11.84NDD730 pKa = 3.06MDD732 pKa = 4.2QSQEE736 pKa = 3.69TWDD739 pKa = 3.79VSGQTPGIYY748 pKa = 10.2DD749 pKa = 3.44AMLPIIEE756 pKa = 4.44TWYY759 pKa = 9.9EE760 pKa = 3.59NYY762 pKa = 10.73GFVGSYY768 pKa = 9.06YY769 pKa = 11.14VNIGSNPPDD778 pKa = 3.53QQTDD782 pKa = 3.02WSVSKK787 pKa = 10.21PYY789 pKa = 10.59YY790 pKa = 10.31DD791 pKa = 3.71RR792 pKa = 11.84LLALEE797 pKa = 4.52SEE799 pKa = 4.8IGTHH803 pKa = 6.29SYY805 pKa = 7.94THH807 pKa = 6.92AVDD810 pKa = 3.73TNLLFPDD817 pKa = 3.96VVTDD821 pKa = 3.66EE822 pKa = 4.77LLQARR827 pKa = 11.84NFVYY831 pKa = 10.63PDD833 pKa = 3.13MTASEE838 pKa = 4.58FNAILADD845 pKa = 3.66TLARR849 pKa = 11.84TDD851 pKa = 3.59PTNPNAIDD859 pKa = 3.91PADD862 pKa = 3.58LTVQEE867 pKa = 4.58KK868 pKa = 10.55AVLEE872 pKa = 3.89ASYY875 pKa = 10.7RR876 pKa = 11.84FQFEE880 pKa = 3.87YY881 pKa = 10.69SKK883 pKa = 11.19LVIEE887 pKa = 4.74RR888 pKa = 11.84EE889 pKa = 3.75MGIQVTGAAVSGAPEE904 pKa = 4.93RR905 pKa = 11.84IDD907 pKa = 3.16ATRR910 pKa = 11.84EE911 pKa = 3.54IIQFFDD917 pKa = 4.39YY918 pKa = 10.8ISGGYY923 pKa = 9.36SGTGAGYY930 pKa = 9.39PGAFGYY936 pKa = 7.39LTPGEE941 pKa = 4.08NDD943 pKa = 3.14RR944 pKa = 11.84VYY946 pKa = 10.8LAPNMSFDD954 pKa = 3.77FSLVGFNGMTPEE966 pKa = 3.88QAEE969 pKa = 4.44ATWLAEE975 pKa = 4.17FAALTANGTTPILAFPWHH993 pKa = 7.57DD994 pKa = 3.74YY995 pKa = 11.49GPTEE999 pKa = 4.38WNLGTPGQVYY1009 pKa = 7.77TFQMYY1014 pKa = 7.68EE1015 pKa = 3.99TLIAAAAASGAEE1027 pKa = 4.16FVTGKK1032 pKa = 10.49DD1033 pKa = 2.97LAEE1036 pKa = 4.95RR1037 pKa = 11.84IASFSASSLTTSRR1050 pKa = 11.84VGDD1053 pKa = 3.71VVNVTVGSSDD1063 pKa = 3.1AGKK1066 pKa = 9.77FAIDD1070 pKa = 4.05LGKK1073 pKa = 10.2EE1074 pKa = 3.98GQIASVTNWYY1084 pKa = 9.98AWDD1087 pKa = 3.75SDD1089 pKa = 4.52SVFLPSSGGTFEE1101 pKa = 4.2ITLGTEE1107 pKa = 3.77MADD1110 pKa = 3.36VTRR1113 pKa = 11.84ISALPMRR1120 pKa = 11.84GDD1122 pKa = 3.81LLSVSGDD1129 pKa = 3.27GSDD1132 pKa = 4.76LDD1134 pKa = 4.28FSFSGRR1140 pKa = 11.84GEE1142 pKa = 4.02VLVTLRR1148 pKa = 11.84SQDD1151 pKa = 3.39SAPIRR1156 pKa = 11.84ITGADD1161 pKa = 3.6SATLPDD1167 pKa = 4.45AGSVSLTFDD1176 pKa = 3.34TVGLNTASVDD1186 pKa = 3.58YY1187 pKa = 10.12LAPGTLLIGDD1197 pKa = 4.69AGSDD1201 pKa = 3.14ILLGGSAADD1210 pKa = 3.64TLEE1213 pKa = 4.0GRR1215 pKa = 11.84GGNDD1219 pKa = 2.68ILSGGGGDD1227 pKa = 3.85DD1228 pKa = 3.35YY1229 pKa = 11.66FVFRR1233 pKa = 11.84LGDD1236 pKa = 3.65AFDD1239 pKa = 4.91TILDD1243 pKa = 4.04FATGIDD1249 pKa = 3.74TLEE1252 pKa = 3.95LSGFGFADD1260 pKa = 4.71AEE1262 pKa = 4.39AAWGAFVDD1270 pKa = 4.0TAEE1273 pKa = 4.24GLEE1276 pKa = 4.23LTFGDD1281 pKa = 4.01TDD1283 pKa = 3.82RR1284 pKa = 11.84LLLAGLNQGDD1294 pKa = 4.75LSSADD1299 pKa = 3.05IVTDD1303 pKa = 3.85PFLFAA1308 pKa = 5.86
MM1 pKa = 7.15PTIDD5 pKa = 4.75GSISDD10 pKa = 4.04WSPDD14 pKa = 3.14ARR16 pKa = 11.84LDD18 pKa = 3.6TAATGTLGYY27 pKa = 9.98GLWGVADD34 pKa = 3.74PDD36 pKa = 3.72YY37 pKa = 10.8FYY39 pKa = 11.2FAITSDD45 pKa = 3.56DD46 pKa = 4.01VIIGEE51 pKa = 4.38NTTLWLDD58 pKa = 3.38TDD60 pKa = 4.52LDD62 pKa = 4.15RR63 pKa = 11.84GTGYY67 pKa = 10.33QIWGWAGGAEE77 pKa = 4.23YY78 pKa = 10.1QVEE81 pKa = 4.08IAADD85 pKa = 3.77GTARR89 pKa = 11.84LYY91 pKa = 11.13SGGPEE96 pKa = 3.79EE97 pKa = 4.18TLVAEE102 pKa = 4.59LEE104 pKa = 4.44AVRR107 pKa = 11.84SADD110 pKa = 3.14GRR112 pKa = 11.84VLEE115 pKa = 4.11FRR117 pKa = 11.84IDD119 pKa = 3.51RR120 pKa = 11.84SLLAGNPDD128 pKa = 3.47AVRR131 pKa = 11.84VYY133 pKa = 11.62ADD135 pKa = 3.52VNDD138 pKa = 4.12SAFLPNSYY146 pKa = 11.09ANTNFIVAEE155 pKa = 4.11PSPSVVTGGKK165 pKa = 7.65TLDD168 pKa = 3.85GNLVDD173 pKa = 3.99WDD175 pKa = 3.6AATRR179 pKa = 11.84LDD181 pKa = 3.95TPATGAAGYY190 pKa = 10.63AFYY193 pKa = 11.34GDD195 pKa = 3.83LQDD198 pKa = 4.49NVFTFAISTDD208 pKa = 3.18GTAIGANTTIWLDD221 pKa = 3.44TDD223 pKa = 3.9RR224 pKa = 11.84DD225 pKa = 3.89TATGYY230 pKa = 8.53QVWGWAVGGEE240 pKa = 4.05YY241 pKa = 10.42NINITADD248 pKa = 3.38GEE250 pKa = 4.19ARR252 pKa = 11.84LYY254 pKa = 10.24TGAAGEE260 pKa = 4.38TFVANLDD267 pKa = 3.43FAYY270 pKa = 10.71NADD273 pKa = 3.52GTVIEE278 pKa = 4.49IALDD282 pKa = 3.45RR283 pKa = 11.84ALVGNPASMRR293 pKa = 11.84VFADD297 pKa = 3.29VNDD300 pKa = 3.96SVFIPNSYY308 pKa = 11.22ANANFIVGSEE318 pKa = 4.01PTPVIGGITLDD329 pKa = 3.78GSVSEE334 pKa = 4.39WTPEE338 pKa = 4.57SILLTQGGYY347 pKa = 6.9QLRR350 pKa = 11.84GEE352 pKa = 4.53FVVDD356 pKa = 3.65AAGDD360 pKa = 3.58QYY362 pKa = 11.94VFAIEE367 pKa = 4.51SDD369 pKa = 3.43GTTIGANTTIWLDD382 pKa = 3.14TDD384 pKa = 4.36LNSTTGYY391 pKa = 10.2QIWGWAGGAEE401 pKa = 4.19YY402 pKa = 10.63NVNITADD409 pKa = 3.62GQARR413 pKa = 11.84LYY415 pKa = 10.83SGAAGGTYY423 pKa = 9.95IADD426 pKa = 3.77LDD428 pKa = 4.35FGFSADD434 pKa = 3.38GTGFEE439 pKa = 4.36VAISKK444 pKa = 10.45DD445 pKa = 4.02LLSGAPDD452 pKa = 3.35QVRR455 pKa = 11.84ILADD459 pKa = 3.3VNDD462 pKa = 4.1AVFLPGDD469 pKa = 3.63YY470 pKa = 10.8SSANLVVGAPLPPPTDD486 pKa = 3.52APEE489 pKa = 3.74SRR491 pKa = 11.84IAIVYY496 pKa = 9.67SATTAANFYY505 pKa = 10.71DD506 pKa = 3.16ITAYY510 pKa = 10.4GQLFMAAQNQAMQAGIPFDD529 pKa = 4.5LLTEE533 pKa = 4.69ADD535 pKa = 4.33LLDD538 pKa = 3.9PTGLAGYY545 pKa = 9.43DD546 pKa = 3.29AIVFPGFSNVQASQAATIAASLTTAAQDD574 pKa = 3.52YY575 pKa = 8.71GVGMIVAGNFLTNDD589 pKa = 3.34EE590 pKa = 4.66TGAALPGNSYY600 pKa = 11.51ARR602 pKa = 11.84MNALLGVTLEE612 pKa = 4.69GFGATQGITLNASNAANPILDD633 pKa = 4.61GYY635 pKa = 9.62TNGQVVGNYY644 pKa = 10.39GNLSYY649 pKa = 11.58LNFTDD654 pKa = 3.72TTGSGQVLFEE664 pKa = 4.12QVVSDD669 pKa = 3.62GTYY672 pKa = 10.28DD673 pKa = 4.07AVIATQTAGRR683 pKa = 11.84NVHH686 pKa = 6.51FATDD690 pKa = 4.23AIMGNNNILGEE701 pKa = 4.47AIDD704 pKa = 5.3WVLQDD709 pKa = 3.75NAPDD713 pKa = 3.31ISLLMTRR720 pKa = 11.84NSSIFYY726 pKa = 10.21SRR728 pKa = 11.84NDD730 pKa = 3.06MDD732 pKa = 4.2QSQEE736 pKa = 3.69TWDD739 pKa = 3.79VSGQTPGIYY748 pKa = 10.2DD749 pKa = 3.44AMLPIIEE756 pKa = 4.44TWYY759 pKa = 9.9EE760 pKa = 3.59NYY762 pKa = 10.73GFVGSYY768 pKa = 9.06YY769 pKa = 11.14VNIGSNPPDD778 pKa = 3.53QQTDD782 pKa = 3.02WSVSKK787 pKa = 10.21PYY789 pKa = 10.59YY790 pKa = 10.31DD791 pKa = 3.71RR792 pKa = 11.84LLALEE797 pKa = 4.52SEE799 pKa = 4.8IGTHH803 pKa = 6.29SYY805 pKa = 7.94THH807 pKa = 6.92AVDD810 pKa = 3.73TNLLFPDD817 pKa = 3.96VVTDD821 pKa = 3.66EE822 pKa = 4.77LLQARR827 pKa = 11.84NFVYY831 pKa = 10.63PDD833 pKa = 3.13MTASEE838 pKa = 4.58FNAILADD845 pKa = 3.66TLARR849 pKa = 11.84TDD851 pKa = 3.59PTNPNAIDD859 pKa = 3.91PADD862 pKa = 3.58LTVQEE867 pKa = 4.58KK868 pKa = 10.55AVLEE872 pKa = 3.89ASYY875 pKa = 10.7RR876 pKa = 11.84FQFEE880 pKa = 3.87YY881 pKa = 10.69SKK883 pKa = 11.19LVIEE887 pKa = 4.74RR888 pKa = 11.84EE889 pKa = 3.75MGIQVTGAAVSGAPEE904 pKa = 4.93RR905 pKa = 11.84IDD907 pKa = 3.16ATRR910 pKa = 11.84EE911 pKa = 3.54IIQFFDD917 pKa = 4.39YY918 pKa = 10.8ISGGYY923 pKa = 9.36SGTGAGYY930 pKa = 9.39PGAFGYY936 pKa = 7.39LTPGEE941 pKa = 4.08NDD943 pKa = 3.14RR944 pKa = 11.84VYY946 pKa = 10.8LAPNMSFDD954 pKa = 3.77FSLVGFNGMTPEE966 pKa = 3.88QAEE969 pKa = 4.44ATWLAEE975 pKa = 4.17FAALTANGTTPILAFPWHH993 pKa = 7.57DD994 pKa = 3.74YY995 pKa = 11.49GPTEE999 pKa = 4.38WNLGTPGQVYY1009 pKa = 7.77TFQMYY1014 pKa = 7.68EE1015 pKa = 3.99TLIAAAAASGAEE1027 pKa = 4.16FVTGKK1032 pKa = 10.49DD1033 pKa = 2.97LAEE1036 pKa = 4.95RR1037 pKa = 11.84IASFSASSLTTSRR1050 pKa = 11.84VGDD1053 pKa = 3.71VVNVTVGSSDD1063 pKa = 3.1AGKK1066 pKa = 9.77FAIDD1070 pKa = 4.05LGKK1073 pKa = 10.2EE1074 pKa = 3.98GQIASVTNWYY1084 pKa = 9.98AWDD1087 pKa = 3.75SDD1089 pKa = 4.52SVFLPSSGGTFEE1101 pKa = 4.2ITLGTEE1107 pKa = 3.77MADD1110 pKa = 3.36VTRR1113 pKa = 11.84ISALPMRR1120 pKa = 11.84GDD1122 pKa = 3.81LLSVSGDD1129 pKa = 3.27GSDD1132 pKa = 4.76LDD1134 pKa = 4.28FSFSGRR1140 pKa = 11.84GEE1142 pKa = 4.02VLVTLRR1148 pKa = 11.84SQDD1151 pKa = 3.39SAPIRR1156 pKa = 11.84ITGADD1161 pKa = 3.6SATLPDD1167 pKa = 4.45AGSVSLTFDD1176 pKa = 3.34TVGLNTASVDD1186 pKa = 3.58YY1187 pKa = 10.12LAPGTLLIGDD1197 pKa = 4.69AGSDD1201 pKa = 3.14ILLGGSAADD1210 pKa = 3.64TLEE1213 pKa = 4.0GRR1215 pKa = 11.84GGNDD1219 pKa = 2.68ILSGGGGDD1227 pKa = 3.85DD1228 pKa = 3.35YY1229 pKa = 11.66FVFRR1233 pKa = 11.84LGDD1236 pKa = 3.65AFDD1239 pKa = 4.91TILDD1243 pKa = 4.04FATGIDD1249 pKa = 3.74TLEE1252 pKa = 3.95LSGFGFADD1260 pKa = 4.71AEE1262 pKa = 4.39AAWGAFVDD1270 pKa = 4.0TAEE1273 pKa = 4.24GLEE1276 pKa = 4.23LTFGDD1281 pKa = 4.01TDD1283 pKa = 3.82RR1284 pKa = 11.84LLLAGLNQGDD1294 pKa = 4.75LSSADD1299 pKa = 3.05IVTDD1303 pKa = 3.85PFLFAA1308 pKa = 5.86
Molecular weight: 138.22 kDa
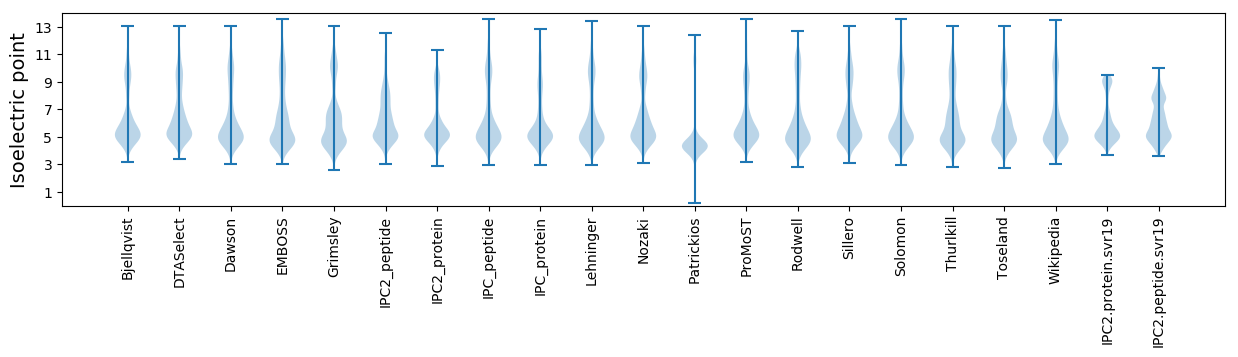
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7JJ68|A0A1H7JJ68_9RHOB Uncharacterized membrane protein YeiH OS=Jannaschia helgolandensis OX=188906 GN=SAMN04488526_1388 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1060539 |
38 |
5064 |
305.5 |
33.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.394 ± 0.057 | 0.844 ± 0.014 |
6.559 ± 0.04 | 5.38 ± 0.038 |
3.569 ± 0.027 | 8.851 ± 0.042 |
1.962 ± 0.022 | 5.252 ± 0.032 |
2.765 ± 0.028 | 10.061 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.836 ± 0.022 | 2.489 ± 0.026 |
5.136 ± 0.039 | 2.996 ± 0.022 |
7.043 ± 0.039 | 5.083 ± 0.03 |
5.903 ± 0.033 | 7.4 ± 0.033 |
1.427 ± 0.019 | 2.049 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
