
Cereal yellow dwarf virus RPS
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 8.34
Get precalculated fractions of proteins
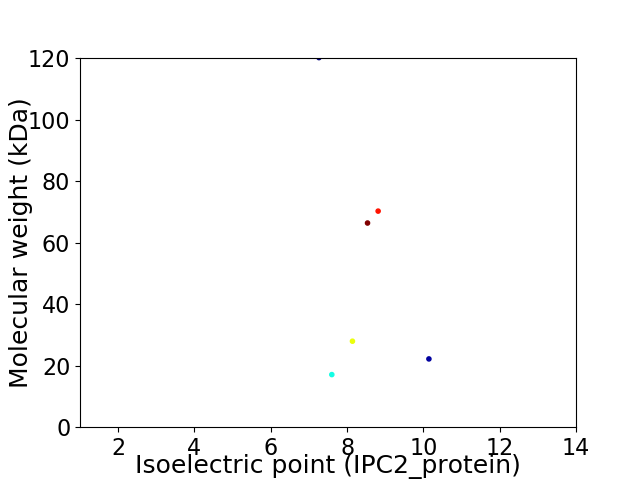
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9IZF6|Q9IZF6_9LUTE Serine protease OS=Cereal yellow dwarf virus RPS OX=228582 PE=4 SV=1
MM1 pKa = 7.45GPKK4 pKa = 9.78KK5 pKa = 10.62LSFSVFLWLMCFLSFSQGNHH25 pKa = 4.7TGMATGSDD33 pKa = 4.08FPSPTTLIYY42 pKa = 10.85SGGVWPLVTPHH53 pKa = 6.71SSPVAGEE60 pKa = 4.24KK61 pKa = 9.43CTLTCAPCPVRR72 pKa = 11.84SITEE76 pKa = 3.73PSYY79 pKa = 11.53KK80 pKa = 10.24EE81 pKa = 4.41LLQQLWVKK89 pKa = 10.62AWLDD93 pKa = 3.46SRR95 pKa = 11.84ALCFTAIDD103 pKa = 3.61SSSRR107 pKa = 11.84FLSHH111 pKa = 7.38AYY113 pKa = 9.4EE114 pKa = 4.38LSSATARR121 pKa = 11.84ALFGQVLWFAVYY133 pKa = 10.07LWTNVLLQTARR144 pKa = 11.84IVCSVVASYY153 pKa = 10.77YY154 pKa = 10.53LQVASLISLGITTSWIYY171 pKa = 10.82KK172 pKa = 7.94ILRR175 pKa = 11.84WTFGTLPASLCIRR188 pKa = 11.84LGKK191 pKa = 9.88SICRR195 pKa = 11.84VLTCRR200 pKa = 11.84KK201 pKa = 9.02FFNEE205 pKa = 3.67KK206 pKa = 10.02SVAGFDD212 pKa = 4.07SYY214 pKa = 11.54SIPQSPPKK222 pKa = 10.26KK223 pKa = 10.28SVITLRR229 pKa = 11.84RR230 pKa = 11.84ADD232 pKa = 3.52KK233 pKa = 9.14THH235 pKa = 6.38VGFAVCIRR243 pKa = 11.84LFNNSNALVTSEE255 pKa = 4.3HH256 pKa = 6.28NLRR259 pKa = 11.84EE260 pKa = 3.73ADD262 pKa = 3.23AFYY265 pKa = 11.08SPRR268 pKa = 11.84TGRR271 pKa = 11.84AIKK274 pKa = 10.18LAEE277 pKa = 4.14FKK279 pKa = 11.12VIFKK283 pKa = 10.77DD284 pKa = 3.1ADD286 pKa = 3.73LDD288 pKa = 3.97VAIVAGPDD296 pKa = 2.87NWEE299 pKa = 4.06SAFGCGSVHH308 pKa = 6.19FTTHH312 pKa = 6.8DD313 pKa = 3.46RR314 pKa = 11.84LAKK317 pKa = 10.27CPAQIYY323 pKa = 10.35VIDD326 pKa = 4.45GEE328 pKa = 4.21DD329 pKa = 3.25WRR331 pKa = 11.84AHH333 pKa = 4.59SAKK336 pKa = 10.47VVGHH340 pKa = 6.66FDD342 pKa = 2.95NFAQVLSNTKK352 pKa = 9.88PGFSGAGYY360 pKa = 9.22FHH362 pKa = 7.39GKK364 pKa = 7.28TLLGVHH370 pKa = 6.77KK371 pKa = 10.32GHH373 pKa = 7.52AGKK376 pKa = 10.24DD377 pKa = 3.49YY378 pKa = 10.82NFNLMAPLPAIPGLTSPKK396 pKa = 10.3YY397 pKa = 9.9EE398 pKa = 4.26IEE400 pKa = 4.06SDD402 pKa = 3.8PPQGLIFPSEE412 pKa = 4.04VAEE415 pKa = 4.86EE416 pKa = 3.97ITKK419 pKa = 9.35TIQSVYY425 pKa = 11.22NDD427 pKa = 3.56FLKK430 pKa = 10.66LDD432 pKa = 3.82KK433 pKa = 10.85SIHH436 pKa = 6.08SKK438 pKa = 9.7GKK440 pKa = 7.18EE441 pKa = 3.84WKK443 pKa = 10.68GGVAWADD450 pKa = 3.59LEE452 pKa = 4.74DD453 pKa = 3.89EE454 pKa = 4.24VRR456 pKa = 11.84KK457 pKa = 10.1VGKK460 pKa = 9.79RR461 pKa = 11.84QGGRR465 pKa = 11.84VRR467 pKa = 11.84RR468 pKa = 11.84TNRR471 pKa = 11.84STKK474 pKa = 9.59SQGRR478 pKa = 11.84YY479 pKa = 8.75KK480 pKa = 10.53RR481 pKa = 11.84NKK483 pKa = 9.29EE484 pKa = 3.97SCCTFFAEE492 pKa = 4.6GTAAPRR498 pKa = 11.84CIPTHH503 pKa = 4.67THH505 pKa = 6.24NLDD508 pKa = 3.98DD509 pKa = 4.07NPMFSRR515 pKa = 11.84CYY517 pKa = 10.56CNIPGHH523 pKa = 6.49KK524 pKa = 10.02CPFWDD529 pKa = 3.61VSARR533 pKa = 11.84RR534 pKa = 11.84DD535 pKa = 3.3AEE537 pKa = 4.09LNEE540 pKa = 4.19FARR543 pKa = 11.84TEE545 pKa = 3.98DD546 pKa = 2.98RR547 pKa = 11.84HH548 pKa = 7.3VEE550 pKa = 3.78DD551 pKa = 5.41RR552 pKa = 11.84EE553 pKa = 3.62IDD555 pKa = 3.68NRR557 pKa = 11.84SGRR560 pKa = 11.84CTSSEE565 pKa = 3.92EE566 pKa = 3.89TSRR569 pKa = 11.84QARR572 pKa = 11.84LQQEE576 pKa = 3.9TRR578 pKa = 11.84AWQQYY583 pKa = 8.62FADD586 pKa = 4.01VYY588 pKa = 8.55TWEE591 pKa = 4.52ISTSEE596 pKa = 3.94QEE598 pKa = 4.06VPGFQRR604 pKa = 11.84VGKK607 pKa = 10.19LSPQYY612 pKa = 10.35YY613 pKa = 9.04PRR615 pKa = 11.84ARR617 pKa = 11.84STTAWGEE624 pKa = 4.12RR625 pKa = 11.84LCAEE629 pKa = 4.48HH630 pKa = 7.31PLLGEE635 pKa = 4.01KK636 pKa = 9.56TKK638 pKa = 11.02GFGWPAVGATAEE650 pKa = 4.28LTSLRR655 pKa = 11.84LQAARR660 pKa = 11.84WLEE663 pKa = 3.8RR664 pKa = 11.84SEE666 pKa = 4.28SAKK669 pKa = 10.39IPSDD673 pKa = 3.28AARR676 pKa = 11.84KK677 pKa = 9.11NVIDD681 pKa = 4.02RR682 pKa = 11.84TVQAYY687 pKa = 8.96SNCKK691 pKa = 9.05TNVPRR696 pKa = 11.84CTRR699 pKa = 11.84DD700 pKa = 3.14QLNWDD705 pKa = 3.97DD706 pKa = 4.37FRR708 pKa = 11.84IDD710 pKa = 3.29FLEE713 pKa = 4.94AIKK716 pKa = 10.62SLQLDD721 pKa = 3.75AGVGIPMITAGLPTHH736 pKa = 6.9RR737 pKa = 11.84GWVEE741 pKa = 4.24DD742 pKa = 4.09PDD744 pKa = 4.35LLPVLARR751 pKa = 11.84LTFDD755 pKa = 4.64RR756 pKa = 11.84LLTMSKK762 pKa = 10.8ASLEE766 pKa = 4.33TRR768 pKa = 11.84SPEE771 pKa = 3.89QLVKK775 pKa = 10.99EE776 pKa = 4.26NLCDD780 pKa = 4.79PIRR783 pKa = 11.84LFVKK787 pKa = 10.15QEE789 pKa = 3.56PHH791 pKa = 6.14KK792 pKa = 10.47QSKK795 pKa = 10.18LDD797 pKa = 3.38EE798 pKa = 4.27GRR800 pKa = 11.84YY801 pKa = 8.96RR802 pKa = 11.84LIMSVSLIDD811 pKa = 3.51QLVARR816 pKa = 11.84VLFQAQNKK824 pKa = 9.79SEE826 pKa = 4.03IALWSAIPSKK836 pKa = 10.64PGFGLSTEE844 pKa = 4.32DD845 pKa = 3.64QVEE848 pKa = 4.23SFINVLADD856 pKa = 3.24TAGARR861 pKa = 11.84PEE863 pKa = 4.31EE864 pKa = 4.68ICDD867 pKa = 3.04KK868 pKa = 10.61WRR870 pKa = 11.84DD871 pKa = 3.63LLVPTDD877 pKa = 3.84CSGFDD882 pKa = 3.34WSVSDD887 pKa = 4.78WMLADD892 pKa = 4.01DD893 pKa = 3.87MEE895 pKa = 4.42VRR897 pKa = 11.84NRR899 pKa = 11.84LTIDD903 pKa = 3.34CNEE906 pKa = 4.17LTRR909 pKa = 11.84HH910 pKa = 5.95LRR912 pKa = 11.84AVWLQGISNSVLCLSDD928 pKa = 3.27GTMLSQVKK936 pKa = 10.09PGVQKK941 pKa = 10.66SGSYY945 pKa = 8.27NTSSTNSRR953 pKa = 11.84IRR955 pKa = 11.84VMAAYY960 pKa = 9.92HH961 pKa = 6.58CGASWAIAMGDD972 pKa = 3.84DD973 pKa = 4.05ALEE976 pKa = 4.56APDD979 pKa = 4.44TDD981 pKa = 3.56LSKK984 pKa = 11.61YY985 pKa = 10.03KK986 pKa = 10.67DD987 pKa = 3.37LGFKK991 pKa = 10.77VEE993 pKa = 4.04VSKK996 pKa = 10.86EE997 pKa = 3.96LEE999 pKa = 4.1FCSHH1003 pKa = 6.51IFKK1006 pKa = 10.88SPTLAIPVNANKK1018 pKa = 9.37MLYY1021 pKa = 10.12RR1022 pKa = 11.84LIHH1025 pKa = 6.62GYY1027 pKa = 9.99NPEE1030 pKa = 4.27CGNAEE1035 pKa = 4.24VIVNYY1040 pKa = 10.48LNAASSVLHH1049 pKa = 6.15EE1050 pKa = 4.61LRR1052 pKa = 11.84HH1053 pKa = 5.37DD1054 pKa = 3.9QEE1056 pKa = 4.61LCALLHH1062 pKa = 5.68MWLVSGITTKK1072 pKa = 11.17DD1073 pKa = 3.01NN1074 pKa = 3.52
MM1 pKa = 7.45GPKK4 pKa = 9.78KK5 pKa = 10.62LSFSVFLWLMCFLSFSQGNHH25 pKa = 4.7TGMATGSDD33 pKa = 4.08FPSPTTLIYY42 pKa = 10.85SGGVWPLVTPHH53 pKa = 6.71SSPVAGEE60 pKa = 4.24KK61 pKa = 9.43CTLTCAPCPVRR72 pKa = 11.84SITEE76 pKa = 3.73PSYY79 pKa = 11.53KK80 pKa = 10.24EE81 pKa = 4.41LLQQLWVKK89 pKa = 10.62AWLDD93 pKa = 3.46SRR95 pKa = 11.84ALCFTAIDD103 pKa = 3.61SSSRR107 pKa = 11.84FLSHH111 pKa = 7.38AYY113 pKa = 9.4EE114 pKa = 4.38LSSATARR121 pKa = 11.84ALFGQVLWFAVYY133 pKa = 10.07LWTNVLLQTARR144 pKa = 11.84IVCSVVASYY153 pKa = 10.77YY154 pKa = 10.53LQVASLISLGITTSWIYY171 pKa = 10.82KK172 pKa = 7.94ILRR175 pKa = 11.84WTFGTLPASLCIRR188 pKa = 11.84LGKK191 pKa = 9.88SICRR195 pKa = 11.84VLTCRR200 pKa = 11.84KK201 pKa = 9.02FFNEE205 pKa = 3.67KK206 pKa = 10.02SVAGFDD212 pKa = 4.07SYY214 pKa = 11.54SIPQSPPKK222 pKa = 10.26KK223 pKa = 10.28SVITLRR229 pKa = 11.84RR230 pKa = 11.84ADD232 pKa = 3.52KK233 pKa = 9.14THH235 pKa = 6.38VGFAVCIRR243 pKa = 11.84LFNNSNALVTSEE255 pKa = 4.3HH256 pKa = 6.28NLRR259 pKa = 11.84EE260 pKa = 3.73ADD262 pKa = 3.23AFYY265 pKa = 11.08SPRR268 pKa = 11.84TGRR271 pKa = 11.84AIKK274 pKa = 10.18LAEE277 pKa = 4.14FKK279 pKa = 11.12VIFKK283 pKa = 10.77DD284 pKa = 3.1ADD286 pKa = 3.73LDD288 pKa = 3.97VAIVAGPDD296 pKa = 2.87NWEE299 pKa = 4.06SAFGCGSVHH308 pKa = 6.19FTTHH312 pKa = 6.8DD313 pKa = 3.46RR314 pKa = 11.84LAKK317 pKa = 10.27CPAQIYY323 pKa = 10.35VIDD326 pKa = 4.45GEE328 pKa = 4.21DD329 pKa = 3.25WRR331 pKa = 11.84AHH333 pKa = 4.59SAKK336 pKa = 10.47VVGHH340 pKa = 6.66FDD342 pKa = 2.95NFAQVLSNTKK352 pKa = 9.88PGFSGAGYY360 pKa = 9.22FHH362 pKa = 7.39GKK364 pKa = 7.28TLLGVHH370 pKa = 6.77KK371 pKa = 10.32GHH373 pKa = 7.52AGKK376 pKa = 10.24DD377 pKa = 3.49YY378 pKa = 10.82NFNLMAPLPAIPGLTSPKK396 pKa = 10.3YY397 pKa = 9.9EE398 pKa = 4.26IEE400 pKa = 4.06SDD402 pKa = 3.8PPQGLIFPSEE412 pKa = 4.04VAEE415 pKa = 4.86EE416 pKa = 3.97ITKK419 pKa = 9.35TIQSVYY425 pKa = 11.22NDD427 pKa = 3.56FLKK430 pKa = 10.66LDD432 pKa = 3.82KK433 pKa = 10.85SIHH436 pKa = 6.08SKK438 pKa = 9.7GKK440 pKa = 7.18EE441 pKa = 3.84WKK443 pKa = 10.68GGVAWADD450 pKa = 3.59LEE452 pKa = 4.74DD453 pKa = 3.89EE454 pKa = 4.24VRR456 pKa = 11.84KK457 pKa = 10.1VGKK460 pKa = 9.79RR461 pKa = 11.84QGGRR465 pKa = 11.84VRR467 pKa = 11.84RR468 pKa = 11.84TNRR471 pKa = 11.84STKK474 pKa = 9.59SQGRR478 pKa = 11.84YY479 pKa = 8.75KK480 pKa = 10.53RR481 pKa = 11.84NKK483 pKa = 9.29EE484 pKa = 3.97SCCTFFAEE492 pKa = 4.6GTAAPRR498 pKa = 11.84CIPTHH503 pKa = 4.67THH505 pKa = 6.24NLDD508 pKa = 3.98DD509 pKa = 4.07NPMFSRR515 pKa = 11.84CYY517 pKa = 10.56CNIPGHH523 pKa = 6.49KK524 pKa = 10.02CPFWDD529 pKa = 3.61VSARR533 pKa = 11.84RR534 pKa = 11.84DD535 pKa = 3.3AEE537 pKa = 4.09LNEE540 pKa = 4.19FARR543 pKa = 11.84TEE545 pKa = 3.98DD546 pKa = 2.98RR547 pKa = 11.84HH548 pKa = 7.3VEE550 pKa = 3.78DD551 pKa = 5.41RR552 pKa = 11.84EE553 pKa = 3.62IDD555 pKa = 3.68NRR557 pKa = 11.84SGRR560 pKa = 11.84CTSSEE565 pKa = 3.92EE566 pKa = 3.89TSRR569 pKa = 11.84QARR572 pKa = 11.84LQQEE576 pKa = 3.9TRR578 pKa = 11.84AWQQYY583 pKa = 8.62FADD586 pKa = 4.01VYY588 pKa = 8.55TWEE591 pKa = 4.52ISTSEE596 pKa = 3.94QEE598 pKa = 4.06VPGFQRR604 pKa = 11.84VGKK607 pKa = 10.19LSPQYY612 pKa = 10.35YY613 pKa = 9.04PRR615 pKa = 11.84ARR617 pKa = 11.84STTAWGEE624 pKa = 4.12RR625 pKa = 11.84LCAEE629 pKa = 4.48HH630 pKa = 7.31PLLGEE635 pKa = 4.01KK636 pKa = 9.56TKK638 pKa = 11.02GFGWPAVGATAEE650 pKa = 4.28LTSLRR655 pKa = 11.84LQAARR660 pKa = 11.84WLEE663 pKa = 3.8RR664 pKa = 11.84SEE666 pKa = 4.28SAKK669 pKa = 10.39IPSDD673 pKa = 3.28AARR676 pKa = 11.84KK677 pKa = 9.11NVIDD681 pKa = 4.02RR682 pKa = 11.84TVQAYY687 pKa = 8.96SNCKK691 pKa = 9.05TNVPRR696 pKa = 11.84CTRR699 pKa = 11.84DD700 pKa = 3.14QLNWDD705 pKa = 3.97DD706 pKa = 4.37FRR708 pKa = 11.84IDD710 pKa = 3.29FLEE713 pKa = 4.94AIKK716 pKa = 10.62SLQLDD721 pKa = 3.75AGVGIPMITAGLPTHH736 pKa = 6.9RR737 pKa = 11.84GWVEE741 pKa = 4.24DD742 pKa = 4.09PDD744 pKa = 4.35LLPVLARR751 pKa = 11.84LTFDD755 pKa = 4.64RR756 pKa = 11.84LLTMSKK762 pKa = 10.8ASLEE766 pKa = 4.33TRR768 pKa = 11.84SPEE771 pKa = 3.89QLVKK775 pKa = 10.99EE776 pKa = 4.26NLCDD780 pKa = 4.79PIRR783 pKa = 11.84LFVKK787 pKa = 10.15QEE789 pKa = 3.56PHH791 pKa = 6.14KK792 pKa = 10.47QSKK795 pKa = 10.18LDD797 pKa = 3.38EE798 pKa = 4.27GRR800 pKa = 11.84YY801 pKa = 8.96RR802 pKa = 11.84LIMSVSLIDD811 pKa = 3.51QLVARR816 pKa = 11.84VLFQAQNKK824 pKa = 9.79SEE826 pKa = 4.03IALWSAIPSKK836 pKa = 10.64PGFGLSTEE844 pKa = 4.32DD845 pKa = 3.64QVEE848 pKa = 4.23SFINVLADD856 pKa = 3.24TAGARR861 pKa = 11.84PEE863 pKa = 4.31EE864 pKa = 4.68ICDD867 pKa = 3.04KK868 pKa = 10.61WRR870 pKa = 11.84DD871 pKa = 3.63LLVPTDD877 pKa = 3.84CSGFDD882 pKa = 3.34WSVSDD887 pKa = 4.78WMLADD892 pKa = 4.01DD893 pKa = 3.87MEE895 pKa = 4.42VRR897 pKa = 11.84NRR899 pKa = 11.84LTIDD903 pKa = 3.34CNEE906 pKa = 4.17LTRR909 pKa = 11.84HH910 pKa = 5.95LRR912 pKa = 11.84AVWLQGISNSVLCLSDD928 pKa = 3.27GTMLSQVKK936 pKa = 10.09PGVQKK941 pKa = 10.66SGSYY945 pKa = 8.27NTSSTNSRR953 pKa = 11.84IRR955 pKa = 11.84VMAAYY960 pKa = 9.92HH961 pKa = 6.58CGASWAIAMGDD972 pKa = 3.84DD973 pKa = 4.05ALEE976 pKa = 4.56APDD979 pKa = 4.44TDD981 pKa = 3.56LSKK984 pKa = 11.61YY985 pKa = 10.03KK986 pKa = 10.67DD987 pKa = 3.37LGFKK991 pKa = 10.77VEE993 pKa = 4.04VSKK996 pKa = 10.86EE997 pKa = 3.96LEE999 pKa = 4.1FCSHH1003 pKa = 6.51IFKK1006 pKa = 10.88SPTLAIPVNANKK1018 pKa = 9.37MLYY1021 pKa = 10.12RR1022 pKa = 11.84LIHH1025 pKa = 6.62GYY1027 pKa = 9.99NPEE1030 pKa = 4.27CGNAEE1035 pKa = 4.24VIVNYY1040 pKa = 10.48LNAASSVLHH1049 pKa = 6.15EE1050 pKa = 4.61LRR1052 pKa = 11.84HH1053 pKa = 5.37DD1054 pKa = 3.9QEE1056 pKa = 4.61LCALLHH1062 pKa = 5.68MWLVSGITTKK1072 pKa = 11.17DD1073 pKa = 3.01NN1074 pKa = 3.52
Molecular weight: 120.21 kDa
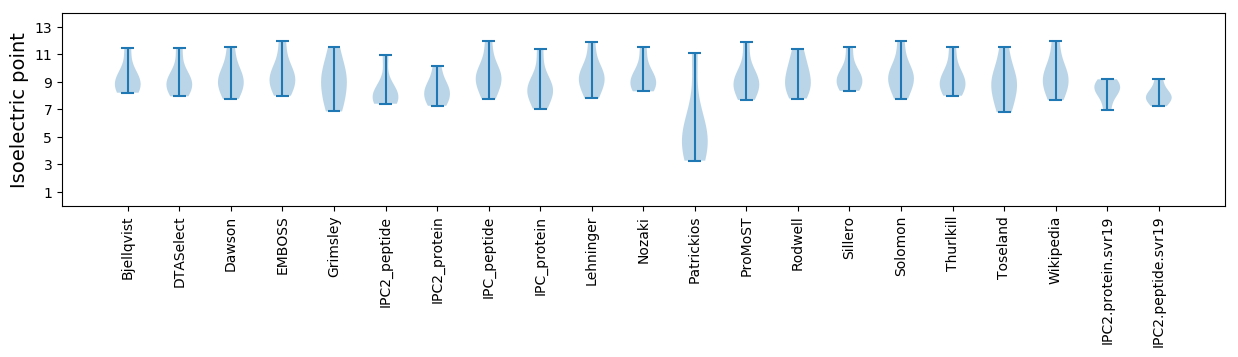
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9IZF4|Q9IZF4_9LUTE Readthrough protein OS=Cereal yellow dwarf virus RPS OX=228582 PE=3 SV=1
MM1 pKa = 6.73STVVLRR7 pKa = 11.84SNGNGSRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84VARR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PAVRR28 pKa = 11.84TQPVVVVTPNGPARR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GPARR51 pKa = 11.84PRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84TPGLGGGGRR65 pKa = 11.84GEE67 pKa = 3.99TFVFTKK73 pKa = 10.68DD74 pKa = 3.19SLAGNSSGSITFGPSLSDD92 pKa = 3.4YY93 pKa = 10.75PAFQNGVLKK102 pKa = 10.51AYY104 pKa = 10.18HH105 pKa = 6.61EE106 pKa = 4.26YY107 pKa = 10.77KK108 pKa = 9.46ITGCILQFVSEE119 pKa = 4.53ASSTAAGSIAYY130 pKa = 9.34EE131 pKa = 4.26LDD133 pKa = 3.41PHH135 pKa = 6.82CKK137 pKa = 9.42ISSLASTNNKK147 pKa = 7.78FTITKK152 pKa = 8.31TGARR156 pKa = 11.84SFPAKK161 pKa = 9.68MINGLEE167 pKa = 3.96WHH169 pKa = 7.13PSDD172 pKa = 4.26EE173 pKa = 4.26DD174 pKa = 3.37QFRR177 pKa = 11.84ILYY180 pKa = 8.98KK181 pKa = 10.99GNGASSVAGSFKK193 pKa = 9.64ITLRR197 pKa = 11.84VQLQNPKK204 pKa = 10.5
MM1 pKa = 6.73STVVLRR7 pKa = 11.84SNGNGSRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84QRR19 pKa = 11.84VARR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84PAVRR28 pKa = 11.84TQPVVVVTPNGPARR42 pKa = 11.84RR43 pKa = 11.84GRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GPARR51 pKa = 11.84PRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84TPGLGGGGRR65 pKa = 11.84GEE67 pKa = 3.99TFVFTKK73 pKa = 10.68DD74 pKa = 3.19SLAGNSSGSITFGPSLSDD92 pKa = 3.4YY93 pKa = 10.75PAFQNGVLKK102 pKa = 10.51AYY104 pKa = 10.18HH105 pKa = 6.61EE106 pKa = 4.26YY107 pKa = 10.77KK108 pKa = 9.46ITGCILQFVSEE119 pKa = 4.53ASSTAAGSIAYY130 pKa = 9.34EE131 pKa = 4.26LDD133 pKa = 3.41PHH135 pKa = 6.82CKK137 pKa = 9.42ISSLASTNNKK147 pKa = 7.78FTITKK152 pKa = 8.31TGARR156 pKa = 11.84SFPAKK161 pKa = 9.68MINGLEE167 pKa = 3.96WHH169 pKa = 7.13PSDD172 pKa = 4.26EE173 pKa = 4.26DD174 pKa = 3.37QFRR177 pKa = 11.84ILYY180 pKa = 8.98KK181 pKa = 10.99GNGASSVAGSFKK193 pKa = 9.64ITLRR197 pKa = 11.84VQLQNPKK204 pKa = 10.5
Molecular weight: 22.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2939 |
153 |
1074 |
489.8 |
54.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.098 ± 0.337 | 1.905 ± 0.513 |
4.491 ± 0.489 | 4.798 ± 0.488 |
3.981 ± 0.306 | 7.043 ± 0.587 |
2.246 ± 0.271 | 4.627 ± 0.186 |
5.104 ± 0.627 | 8.302 ± 1.088 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.327 ± 0.176 | 3.641 ± 0.461 |
6.363 ± 0.776 | 3.879 ± 0.439 |
6.635 ± 0.869 | 10.174 ± 0.751 |
6.227 ± 0.257 | 6.397 ± 0.111 |
1.871 ± 0.297 | 2.858 ± 0.407 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
