
Sewage-associated circular DNA virus-3
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.47
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IZN1|A0A075IZN1_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-3 OX=1519392 PE=4 SV=1
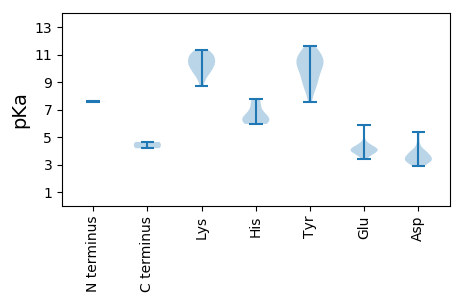
MM1 pKa = 7.59VYY3 pKa = 9.32TFSFTYY9 pKa = 8.33PQCSLEE15 pKa = 4.04RR16 pKa = 11.84TEE18 pKa = 5.91LRR20 pKa = 11.84DD21 pKa = 3.97SLIQRR26 pKa = 11.84VNPEE30 pKa = 3.68KK31 pKa = 10.88YY32 pKa = 10.29LIARR36 pKa = 11.84EE37 pKa = 3.94QHH39 pKa = 6.07SDD41 pKa = 3.23GGLHH45 pKa = 5.95LHH47 pKa = 7.3AYY49 pKa = 9.54LHH51 pKa = 6.2FGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84FTSADD62 pKa = 3.26AFDD65 pKa = 4.01VDD67 pKa = 4.45GFHH70 pKa = 7.75PNIQKK75 pKa = 9.72PRR77 pKa = 11.84SARR80 pKa = 11.84NVIAYY85 pKa = 8.93CSKK88 pKa = 10.99EE89 pKa = 4.03DD90 pKa = 3.97TEE92 pKa = 4.28PMANFDD98 pKa = 4.05YY99 pKa = 11.08TSGEE103 pKa = 4.2PNGGWSEE110 pKa = 3.93ILGRR114 pKa = 11.84TSSKK118 pKa = 11.33DD119 pKa = 3.21EE120 pKa = 3.96FLEE123 pKa = 4.05EE124 pKa = 4.21VRR126 pKa = 11.84THH128 pKa = 6.63FPRR131 pKa = 11.84DD132 pKa = 3.42YY133 pKa = 10.89VLSLEE138 pKa = 4.2RR139 pKa = 11.84LLFFCEE145 pKa = 3.68WRR147 pKa = 11.84FGRR150 pKa = 11.84DD151 pKa = 2.88EE152 pKa = 4.08TEE154 pKa = 3.71YY155 pKa = 10.03TGRR158 pKa = 11.84SRR160 pKa = 11.84TEE162 pKa = 3.38FRR164 pKa = 11.84EE165 pKa = 3.8PDD167 pKa = 3.43TLTNWVNTNLLQVCIWTYY185 pKa = 10.35GPRR188 pKa = 11.84GGPQSPPLLSSPCLYY203 pKa = 10.12WLIWTQWARR212 pKa = 11.84SIDD215 pKa = 3.85STHH218 pKa = 6.41VYY220 pKa = 8.49CQGMFNLDD228 pKa = 3.14EE229 pKa = 4.66WNDD232 pKa = 3.32KK233 pKa = 10.65AKK235 pKa = 10.91YY236 pKa = 10.5VIFDD240 pKa = 5.36DD241 pKa = 4.71IDD243 pKa = 3.31IKK245 pKa = 11.17YY246 pKa = 9.25FPHH249 pKa = 6.34WKK251 pKa = 10.19SILGCQRR258 pKa = 11.84DD259 pKa = 3.49IQLTDD264 pKa = 2.92KK265 pKa = 10.61YY266 pKa = 10.35RR267 pKa = 11.84KK268 pKa = 9.41KK269 pKa = 10.33RR270 pKa = 11.84RR271 pKa = 11.84LRR273 pKa = 11.84NGLPCVWLCNEE284 pKa = 4.37DD285 pKa = 3.42MDD287 pKa = 5.28PRR289 pKa = 11.84GALSRR294 pKa = 11.84TQCEE298 pKa = 4.46WIEE301 pKa = 4.48SNCDD305 pKa = 3.11VVTLRR310 pKa = 11.84EE311 pKa = 4.1PLFEE315 pKa = 4.19
MM1 pKa = 7.59VYY3 pKa = 9.32TFSFTYY9 pKa = 8.33PQCSLEE15 pKa = 4.04RR16 pKa = 11.84TEE18 pKa = 5.91LRR20 pKa = 11.84DD21 pKa = 3.97SLIQRR26 pKa = 11.84VNPEE30 pKa = 3.68KK31 pKa = 10.88YY32 pKa = 10.29LIARR36 pKa = 11.84EE37 pKa = 3.94QHH39 pKa = 6.07SDD41 pKa = 3.23GGLHH45 pKa = 5.95LHH47 pKa = 7.3AYY49 pKa = 9.54LHH51 pKa = 6.2FGRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84RR57 pKa = 11.84FTSADD62 pKa = 3.26AFDD65 pKa = 4.01VDD67 pKa = 4.45GFHH70 pKa = 7.75PNIQKK75 pKa = 9.72PRR77 pKa = 11.84SARR80 pKa = 11.84NVIAYY85 pKa = 8.93CSKK88 pKa = 10.99EE89 pKa = 4.03DD90 pKa = 3.97TEE92 pKa = 4.28PMANFDD98 pKa = 4.05YY99 pKa = 11.08TSGEE103 pKa = 4.2PNGGWSEE110 pKa = 3.93ILGRR114 pKa = 11.84TSSKK118 pKa = 11.33DD119 pKa = 3.21EE120 pKa = 3.96FLEE123 pKa = 4.05EE124 pKa = 4.21VRR126 pKa = 11.84THH128 pKa = 6.63FPRR131 pKa = 11.84DD132 pKa = 3.42YY133 pKa = 10.89VLSLEE138 pKa = 4.2RR139 pKa = 11.84LLFFCEE145 pKa = 3.68WRR147 pKa = 11.84FGRR150 pKa = 11.84DD151 pKa = 2.88EE152 pKa = 4.08TEE154 pKa = 3.71YY155 pKa = 10.03TGRR158 pKa = 11.84SRR160 pKa = 11.84TEE162 pKa = 3.38FRR164 pKa = 11.84EE165 pKa = 3.8PDD167 pKa = 3.43TLTNWVNTNLLQVCIWTYY185 pKa = 10.35GPRR188 pKa = 11.84GGPQSPPLLSSPCLYY203 pKa = 10.12WLIWTQWARR212 pKa = 11.84SIDD215 pKa = 3.85STHH218 pKa = 6.41VYY220 pKa = 8.49CQGMFNLDD228 pKa = 3.14EE229 pKa = 4.66WNDD232 pKa = 3.32KK233 pKa = 10.65AKK235 pKa = 10.91YY236 pKa = 10.5VIFDD240 pKa = 5.36DD241 pKa = 4.71IDD243 pKa = 3.31IKK245 pKa = 11.17YY246 pKa = 9.25FPHH249 pKa = 6.34WKK251 pKa = 10.19SILGCQRR258 pKa = 11.84DD259 pKa = 3.49IQLTDD264 pKa = 2.92KK265 pKa = 10.61YY266 pKa = 10.35RR267 pKa = 11.84KK268 pKa = 9.41KK269 pKa = 10.33RR270 pKa = 11.84RR271 pKa = 11.84LRR273 pKa = 11.84NGLPCVWLCNEE284 pKa = 4.37DD285 pKa = 3.42MDD287 pKa = 5.28PRR289 pKa = 11.84GALSRR294 pKa = 11.84TQCEE298 pKa = 4.46WIEE301 pKa = 4.48SNCDD305 pKa = 3.11VVTLRR310 pKa = 11.84EE311 pKa = 4.1PLFEE315 pKa = 4.19
Molecular weight: 37.28 kDa
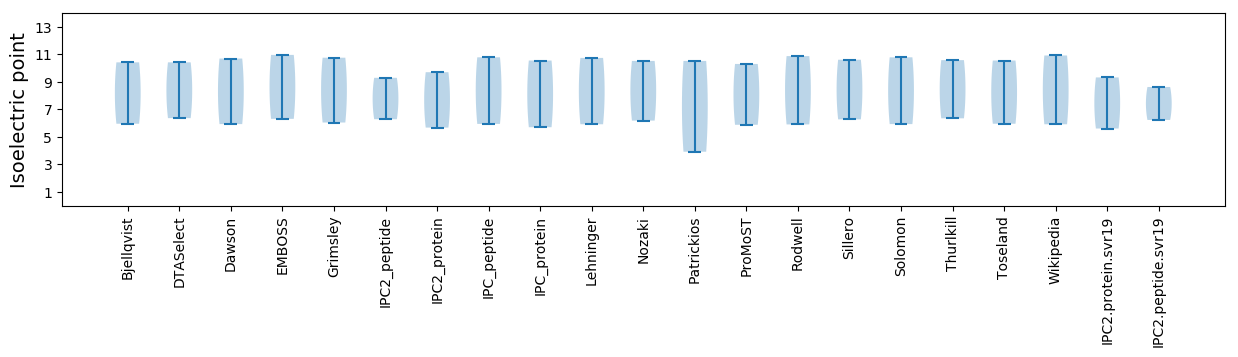
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075IZN1|A0A075IZN1_9VIRU Putative capsid protein OS=Sewage-associated circular DNA virus-3 OX=1519392 PE=4 SV=1
MM1 pKa = 7.58ASSLGQLAFGLAATAAVYY19 pKa = 9.99PLTRR23 pKa = 11.84EE24 pKa = 3.42VDD26 pKa = 3.46NYY28 pKa = 10.84VSNKK32 pKa = 9.65RR33 pKa = 11.84KK34 pKa = 10.35SEE36 pKa = 3.96DD37 pKa = 3.23SRR39 pKa = 11.84SFSVKK44 pKa = 9.87RR45 pKa = 11.84LKK47 pKa = 10.99LSTNKK52 pKa = 10.33DD53 pKa = 2.88MAFRR57 pKa = 11.84SRR59 pKa = 11.84RR60 pKa = 11.84SFRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84PRR67 pKa = 11.84RR68 pKa = 11.84FRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84FRR74 pKa = 11.84KK75 pKa = 9.14RR76 pKa = 11.84RR77 pKa = 11.84MPFRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84FTRR87 pKa = 11.84AVRR90 pKa = 11.84RR91 pKa = 11.84VILRR95 pKa = 11.84TSEE98 pKa = 4.18PKK100 pKa = 10.3QVWAPEE106 pKa = 4.0NVTGYY111 pKa = 10.9AIRR114 pKa = 11.84EE115 pKa = 3.92GDD117 pKa = 3.38GTSRR121 pKa = 11.84VINVSNPPAAPIQGVEE137 pKa = 3.87DD138 pKa = 3.73DD139 pKa = 3.9QYY141 pKa = 11.62IGNKK145 pKa = 9.72YY146 pKa = 8.75FLKK149 pKa = 10.86GIALRR154 pKa = 11.84GQVGTSGEE162 pKa = 4.11VTNRR166 pKa = 11.84QGCLIRR172 pKa = 11.84VTLVWSKK179 pKa = 10.51EE180 pKa = 3.71QAPNLFGAWTEE191 pKa = 4.22FTSATTSTAAPLQTPPDD208 pKa = 3.7LNPRR212 pKa = 11.84FFAITNGTQEE222 pKa = 4.02FVGNGWAIPFDD233 pKa = 3.61RR234 pKa = 11.84TRR236 pKa = 11.84VRR238 pKa = 11.84VISSKK243 pKa = 9.97TIVVNPGVDD252 pKa = 3.31NEE254 pKa = 4.38AGAGIISMPTAFSLYY269 pKa = 10.13FKK271 pKa = 10.18VNKK274 pKa = 8.7WMQIEE279 pKa = 5.17DD280 pKa = 3.78NDD282 pKa = 3.81QSDD285 pKa = 3.85MSSSLISFKK294 pKa = 11.11YY295 pKa = 7.53GTYY298 pKa = 9.32YY299 pKa = 11.35LVMQAVSNTNDD310 pKa = 3.15VTDD313 pKa = 3.55QTIAEE318 pKa = 4.23MDD320 pKa = 3.45YY321 pKa = 11.09SIGVFYY327 pKa = 10.72RR328 pKa = 11.84DD329 pKa = 3.36PP330 pKa = 4.65
MM1 pKa = 7.58ASSLGQLAFGLAATAAVYY19 pKa = 9.99PLTRR23 pKa = 11.84EE24 pKa = 3.42VDD26 pKa = 3.46NYY28 pKa = 10.84VSNKK32 pKa = 9.65RR33 pKa = 11.84KK34 pKa = 10.35SEE36 pKa = 3.96DD37 pKa = 3.23SRR39 pKa = 11.84SFSVKK44 pKa = 9.87RR45 pKa = 11.84LKK47 pKa = 10.99LSTNKK52 pKa = 10.33DD53 pKa = 2.88MAFRR57 pKa = 11.84SRR59 pKa = 11.84RR60 pKa = 11.84SFRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84PRR67 pKa = 11.84RR68 pKa = 11.84FRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84FRR74 pKa = 11.84KK75 pKa = 9.14RR76 pKa = 11.84RR77 pKa = 11.84MPFRR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84FTRR87 pKa = 11.84AVRR90 pKa = 11.84RR91 pKa = 11.84VILRR95 pKa = 11.84TSEE98 pKa = 4.18PKK100 pKa = 10.3QVWAPEE106 pKa = 4.0NVTGYY111 pKa = 10.9AIRR114 pKa = 11.84EE115 pKa = 3.92GDD117 pKa = 3.38GTSRR121 pKa = 11.84VINVSNPPAAPIQGVEE137 pKa = 3.87DD138 pKa = 3.73DD139 pKa = 3.9QYY141 pKa = 11.62IGNKK145 pKa = 9.72YY146 pKa = 8.75FLKK149 pKa = 10.86GIALRR154 pKa = 11.84GQVGTSGEE162 pKa = 4.11VTNRR166 pKa = 11.84QGCLIRR172 pKa = 11.84VTLVWSKK179 pKa = 10.51EE180 pKa = 3.71QAPNLFGAWTEE191 pKa = 4.22FTSATTSTAAPLQTPPDD208 pKa = 3.7LNPRR212 pKa = 11.84FFAITNGTQEE222 pKa = 4.02FVGNGWAIPFDD233 pKa = 3.61RR234 pKa = 11.84TRR236 pKa = 11.84VRR238 pKa = 11.84VISSKK243 pKa = 9.97TIVVNPGVDD252 pKa = 3.31NEE254 pKa = 4.38AGAGIISMPTAFSLYY269 pKa = 10.13FKK271 pKa = 10.18VNKK274 pKa = 8.7WMQIEE279 pKa = 5.17DD280 pKa = 3.78NDD282 pKa = 3.81QSDD285 pKa = 3.85MSSSLISFKK294 pKa = 11.11YY295 pKa = 7.53GTYY298 pKa = 9.32YY299 pKa = 11.35LVMQAVSNTNDD310 pKa = 3.15VTDD313 pKa = 3.55QTIAEE318 pKa = 4.23MDD320 pKa = 3.45YY321 pKa = 11.09SIGVFYY327 pKa = 10.72RR328 pKa = 11.84DD329 pKa = 3.36PP330 pKa = 4.65
Molecular weight: 37.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
645 |
315 |
330 |
322.5 |
37.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.426 ± 1.583 | 1.86 ± 1.147 |
5.891 ± 0.768 | 5.581 ± 1.21 |
5.581 ± 0.13 | 5.891 ± 0.348 |
1.24 ± 0.914 | 4.806 ± 0.254 |
3.876 ± 0.27 | 7.132 ± 1.459 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.415 | 4.806 ± 0.478 |
5.271 ± 0.088 | 3.721 ± 0.161 |
10.078 ± 0.613 | 7.597 ± 0.654 |
6.977 ± 0.441 | 6.047 ± 1.35 |
2.481 ± 0.711 | 3.876 ± 0.4 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
