
Equus asinus papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyochipapillomavirus; Dyochipapillomavirus 1
Average proteome isoelectric point is 5.85
Get precalculated fractions of proteins
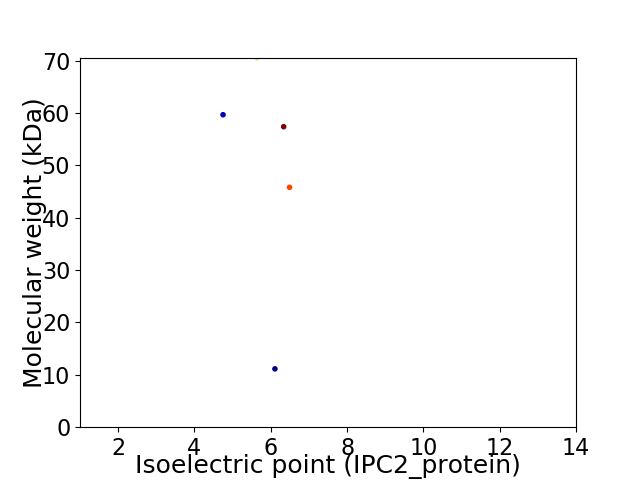
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5ZT83|W5ZT83_9PAPI Regulatory protein E2 OS=Equus asinus papillomavirus 1 OX=1163703 GN=E2 PE=3 SV=1
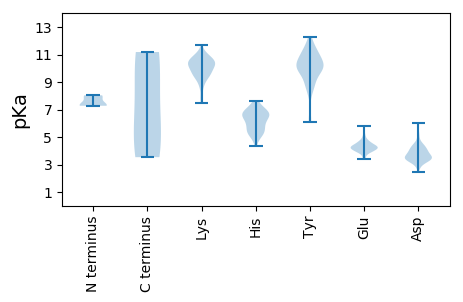
MM1 pKa = 7.95PILARR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 9.0RR10 pKa = 11.84RR11 pKa = 11.84AAPQDD16 pKa = 3.94LYY18 pKa = 10.47PACKK22 pKa = 9.38QANTCPPDD30 pKa = 3.52VLAKK34 pKa = 9.95MEE36 pKa = 4.51NDD38 pKa = 2.89TWADD42 pKa = 4.33RR43 pKa = 11.84ILKK46 pKa = 7.95WASGFLYY53 pKa = 10.39FGNLGISTGRR63 pKa = 11.84GTGGGFGYY71 pKa = 10.92APIGGSGVATGGRR84 pKa = 11.84TVVRR88 pKa = 11.84PGIGANVVVPGEE100 pKa = 4.03AVPTEE105 pKa = 4.18TLGPLDD111 pKa = 4.37PSVIPLDD118 pKa = 3.68TLPSVEE124 pKa = 4.88PDD126 pKa = 3.1VLLPPGRR133 pKa = 11.84PEE135 pKa = 3.86VYY137 pKa = 9.77EE138 pKa = 4.66DD139 pKa = 3.53SLGPRR144 pKa = 11.84IPPEE148 pKa = 4.26RR149 pKa = 11.84LPAAYY154 pKa = 9.97DD155 pKa = 3.53EE156 pKa = 5.57DD157 pKa = 4.63GPPVHH162 pKa = 6.52VPAVPADD169 pKa = 3.77PAVHH173 pKa = 6.63EE174 pKa = 4.45GTPAVLEE181 pKa = 4.5VPTEE185 pKa = 3.88QAIVSHH191 pKa = 5.79SQFANPAFEE200 pKa = 4.43ASIVDD205 pKa = 3.25ITGAADD211 pKa = 3.03ISGQQSITVHH221 pKa = 6.33HH222 pKa = 6.52GAEE225 pKa = 4.02GHH227 pKa = 6.75IIGGDD232 pKa = 3.42VLINPFLEE240 pKa = 4.28STEE243 pKa = 4.18TLEE246 pKa = 5.12GEE248 pKa = 4.57HH249 pKa = 6.47IPLRR253 pKa = 11.84SFTSSMRR260 pKa = 11.84DD261 pKa = 2.93EE262 pKa = 4.38FEE264 pKa = 4.49YY265 pKa = 11.38NEE267 pKa = 4.1EE268 pKa = 4.26TPLQSTPTEE277 pKa = 4.09RR278 pKa = 11.84PAPRR282 pKa = 11.84GRR284 pKa = 11.84RR285 pKa = 11.84LGNTRR290 pKa = 11.84YY291 pKa = 8.79FRR293 pKa = 11.84QVKK296 pKa = 9.75VEE298 pKa = 3.93DD299 pKa = 3.94TLFLHH304 pKa = 6.81HH305 pKa = 6.77PQRR308 pKa = 11.84LVTFDD313 pKa = 3.34NPVYY317 pKa = 10.32EE318 pKa = 5.07GEE320 pKa = 4.14DD321 pKa = 3.28SYY323 pKa = 12.25GFAEE327 pKa = 4.86ALDD330 pKa = 4.22PSAPYY335 pKa = 9.85PDD337 pKa = 5.15LDD339 pKa = 4.03FRR341 pKa = 11.84DD342 pKa = 4.05LKK344 pKa = 10.9SLSRR348 pKa = 11.84VEE350 pKa = 4.16YY351 pKa = 10.43GRR353 pKa = 11.84NPQGLVRR360 pKa = 11.84VSRR363 pKa = 11.84VGKK366 pKa = 10.15AHH368 pKa = 5.38GTMLRR373 pKa = 11.84SGLRR377 pKa = 11.84IGPEE381 pKa = 3.39THH383 pKa = 6.22YY384 pKa = 11.2FMDD387 pKa = 4.06LSEE390 pKa = 4.37IAVEE394 pKa = 4.61SEE396 pKa = 4.06LSLHH400 pKa = 6.81AEE402 pKa = 4.16TSHH405 pKa = 5.46MVGRR409 pKa = 11.84EE410 pKa = 3.93AAQVTPLMEE419 pKa = 5.14EE420 pKa = 3.44ISLISEE426 pKa = 4.39AGSVYY431 pKa = 10.61SDD433 pKa = 3.54SDD435 pKa = 3.98LLDD438 pKa = 4.01VIEE441 pKa = 5.25DD442 pKa = 3.47LGEE445 pKa = 4.09RR446 pKa = 11.84LQLVIGSGRR455 pKa = 11.84NRR457 pKa = 11.84LQVPLPLRR465 pKa = 11.84NLSYY469 pKa = 11.26LDD471 pKa = 3.25TSGIFVGYY479 pKa = 9.01VIPTEE484 pKa = 4.6GQDD487 pKa = 3.04STQRR491 pKa = 11.84SDD493 pKa = 3.32IEE495 pKa = 4.22DD496 pKa = 3.38VSEE499 pKa = 4.28VFSAADD505 pKa = 3.97LSPSFDD511 pKa = 3.28WPMTDD516 pKa = 4.07LDD518 pKa = 4.57ADD520 pKa = 4.4FVLHH524 pKa = 6.74PSLLKK529 pKa = 10.06RR530 pKa = 11.84RR531 pKa = 11.84RR532 pKa = 11.84RR533 pKa = 11.84FYY535 pKa = 10.58WSFADD540 pKa = 4.07GGLASRR546 pKa = 11.84TKK548 pKa = 10.81
MM1 pKa = 7.95PILARR6 pKa = 11.84SRR8 pKa = 11.84KK9 pKa = 9.0RR10 pKa = 11.84RR11 pKa = 11.84AAPQDD16 pKa = 3.94LYY18 pKa = 10.47PACKK22 pKa = 9.38QANTCPPDD30 pKa = 3.52VLAKK34 pKa = 9.95MEE36 pKa = 4.51NDD38 pKa = 2.89TWADD42 pKa = 4.33RR43 pKa = 11.84ILKK46 pKa = 7.95WASGFLYY53 pKa = 10.39FGNLGISTGRR63 pKa = 11.84GTGGGFGYY71 pKa = 10.92APIGGSGVATGGRR84 pKa = 11.84TVVRR88 pKa = 11.84PGIGANVVVPGEE100 pKa = 4.03AVPTEE105 pKa = 4.18TLGPLDD111 pKa = 4.37PSVIPLDD118 pKa = 3.68TLPSVEE124 pKa = 4.88PDD126 pKa = 3.1VLLPPGRR133 pKa = 11.84PEE135 pKa = 3.86VYY137 pKa = 9.77EE138 pKa = 4.66DD139 pKa = 3.53SLGPRR144 pKa = 11.84IPPEE148 pKa = 4.26RR149 pKa = 11.84LPAAYY154 pKa = 9.97DD155 pKa = 3.53EE156 pKa = 5.57DD157 pKa = 4.63GPPVHH162 pKa = 6.52VPAVPADD169 pKa = 3.77PAVHH173 pKa = 6.63EE174 pKa = 4.45GTPAVLEE181 pKa = 4.5VPTEE185 pKa = 3.88QAIVSHH191 pKa = 5.79SQFANPAFEE200 pKa = 4.43ASIVDD205 pKa = 3.25ITGAADD211 pKa = 3.03ISGQQSITVHH221 pKa = 6.33HH222 pKa = 6.52GAEE225 pKa = 4.02GHH227 pKa = 6.75IIGGDD232 pKa = 3.42VLINPFLEE240 pKa = 4.28STEE243 pKa = 4.18TLEE246 pKa = 5.12GEE248 pKa = 4.57HH249 pKa = 6.47IPLRR253 pKa = 11.84SFTSSMRR260 pKa = 11.84DD261 pKa = 2.93EE262 pKa = 4.38FEE264 pKa = 4.49YY265 pKa = 11.38NEE267 pKa = 4.1EE268 pKa = 4.26TPLQSTPTEE277 pKa = 4.09RR278 pKa = 11.84PAPRR282 pKa = 11.84GRR284 pKa = 11.84RR285 pKa = 11.84LGNTRR290 pKa = 11.84YY291 pKa = 8.79FRR293 pKa = 11.84QVKK296 pKa = 9.75VEE298 pKa = 3.93DD299 pKa = 3.94TLFLHH304 pKa = 6.81HH305 pKa = 6.77PQRR308 pKa = 11.84LVTFDD313 pKa = 3.34NPVYY317 pKa = 10.32EE318 pKa = 5.07GEE320 pKa = 4.14DD321 pKa = 3.28SYY323 pKa = 12.25GFAEE327 pKa = 4.86ALDD330 pKa = 4.22PSAPYY335 pKa = 9.85PDD337 pKa = 5.15LDD339 pKa = 4.03FRR341 pKa = 11.84DD342 pKa = 4.05LKK344 pKa = 10.9SLSRR348 pKa = 11.84VEE350 pKa = 4.16YY351 pKa = 10.43GRR353 pKa = 11.84NPQGLVRR360 pKa = 11.84VSRR363 pKa = 11.84VGKK366 pKa = 10.15AHH368 pKa = 5.38GTMLRR373 pKa = 11.84SGLRR377 pKa = 11.84IGPEE381 pKa = 3.39THH383 pKa = 6.22YY384 pKa = 11.2FMDD387 pKa = 4.06LSEE390 pKa = 4.37IAVEE394 pKa = 4.61SEE396 pKa = 4.06LSLHH400 pKa = 6.81AEE402 pKa = 4.16TSHH405 pKa = 5.46MVGRR409 pKa = 11.84EE410 pKa = 3.93AAQVTPLMEE419 pKa = 5.14EE420 pKa = 3.44ISLISEE426 pKa = 4.39AGSVYY431 pKa = 10.61SDD433 pKa = 3.54SDD435 pKa = 3.98LLDD438 pKa = 4.01VIEE441 pKa = 5.25DD442 pKa = 3.47LGEE445 pKa = 4.09RR446 pKa = 11.84LQLVIGSGRR455 pKa = 11.84NRR457 pKa = 11.84LQVPLPLRR465 pKa = 11.84NLSYY469 pKa = 11.26LDD471 pKa = 3.25TSGIFVGYY479 pKa = 9.01VIPTEE484 pKa = 4.6GQDD487 pKa = 3.04STQRR491 pKa = 11.84SDD493 pKa = 3.32IEE495 pKa = 4.22DD496 pKa = 3.38VSEE499 pKa = 4.28VFSAADD505 pKa = 3.97LSPSFDD511 pKa = 3.28WPMTDD516 pKa = 4.07LDD518 pKa = 4.57ADD520 pKa = 4.4FVLHH524 pKa = 6.74PSLLKK529 pKa = 10.06RR530 pKa = 11.84RR531 pKa = 11.84RR532 pKa = 11.84RR533 pKa = 11.84FYY535 pKa = 10.58WSFADD540 pKa = 4.07GGLASRR546 pKa = 11.84TKK548 pKa = 10.81
Molecular weight: 59.66 kDa
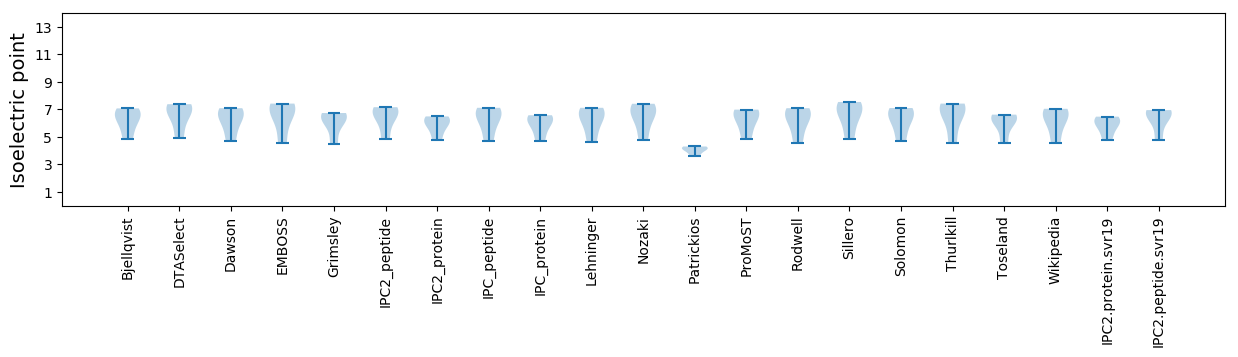
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W6A1G2|W6A1G2_9PAPI Replication protein E1 OS=Equus asinus papillomavirus 1 OX=1163703 GN=E1 PE=3 SV=1
MM1 pKa = 7.27TALQEE6 pKa = 4.34RR7 pKa = 11.84LDD9 pKa = 4.08SVQEE13 pKa = 3.84QQLQIIEE20 pKa = 4.3TGTTKK25 pKa = 10.75LDD27 pKa = 3.68DD28 pKa = 4.84LLVFWRR34 pKa = 11.84SVRR37 pKa = 11.84QEE39 pKa = 3.37NALLHH44 pKa = 6.05FARR47 pKa = 11.84QKK49 pKa = 10.05GLSRR53 pKa = 11.84IDD55 pKa = 3.58LTPVPSLAASACKK68 pKa = 9.89AKK70 pKa = 10.31QAIEE74 pKa = 4.2IEE76 pKa = 4.24LSLEE80 pKa = 4.39GIKK83 pKa = 10.84NSGWDD88 pKa = 3.71DD89 pKa = 4.21EE90 pKa = 4.67PWTLQDD96 pKa = 4.35GSWDD100 pKa = 3.58RR101 pKa = 11.84YY102 pKa = 9.01AASPEE107 pKa = 4.03GAFKK111 pKa = 10.94KK112 pKa = 10.55NPAVVEE118 pKa = 4.55VIFDD122 pKa = 4.08EE123 pKa = 4.47NPEE126 pKa = 3.87NAVWYY131 pKa = 6.2THH133 pKa = 6.42WEE135 pKa = 3.98VLYY138 pKa = 9.9VTDD141 pKa = 3.94GVKK144 pKa = 8.75WHH146 pKa = 6.55RR147 pKa = 11.84TTGRR151 pKa = 11.84VCANGLFYY159 pKa = 10.78EE160 pKa = 4.77LGGEE164 pKa = 3.88QTFYY168 pKa = 11.52EE169 pKa = 4.72RR170 pKa = 11.84FDD172 pKa = 4.54LDD174 pKa = 3.37AQKK177 pKa = 11.16YY178 pKa = 6.09STGGTWTVTFQGEE191 pKa = 4.66TMTGSLVSSSTDD203 pKa = 3.25GVDD206 pKa = 3.59GEE208 pKa = 4.79ARR210 pKa = 11.84PPSPMPASTACGEE223 pKa = 4.1RR224 pKa = 11.84RR225 pKa = 11.84DD226 pKa = 4.15NLSTPAEE233 pKa = 4.41PDD235 pKa = 2.94SSSAVSSNFTSSGGRR250 pKa = 11.84SRR252 pKa = 11.84GRR254 pKa = 11.84ANRR257 pKa = 11.84GCPYY261 pKa = 10.09TRR263 pKa = 11.84TEE265 pKa = 4.21GPRR268 pKa = 11.84VPKK271 pKa = 10.44VVSAPCYY278 pKa = 9.69RR279 pKa = 11.84PRR281 pKa = 11.84GVQAQLGSTPPLVSPADD298 pKa = 3.57PLGRR302 pKa = 11.84SSAGEE307 pKa = 4.17RR308 pKa = 11.84PPGAAGHH315 pKa = 6.17SGSVGNAVPPAAPPASPFLQGSRR338 pKa = 11.84CPVILVTGSCNQVKK352 pKa = 9.74CWRR355 pKa = 11.84LRR357 pKa = 11.84CRR359 pKa = 11.84RR360 pKa = 11.84HH361 pKa = 5.06HH362 pKa = 5.27RR363 pKa = 11.84HH364 pKa = 5.93RR365 pKa = 11.84YY366 pKa = 8.44HH367 pKa = 7.2RR368 pKa = 11.84VTSTWYY374 pKa = 8.43ITGDD378 pKa = 3.38QGNDD382 pKa = 2.83RR383 pKa = 11.84AGPSTVMVMFTDD395 pKa = 4.24DD396 pKa = 3.26AQRR399 pKa = 11.84ADD401 pKa = 4.05FMAKK405 pKa = 9.6VSFPPGVSCTPLTLSTT421 pKa = 4.27
MM1 pKa = 7.27TALQEE6 pKa = 4.34RR7 pKa = 11.84LDD9 pKa = 4.08SVQEE13 pKa = 3.84QQLQIIEE20 pKa = 4.3TGTTKK25 pKa = 10.75LDD27 pKa = 3.68DD28 pKa = 4.84LLVFWRR34 pKa = 11.84SVRR37 pKa = 11.84QEE39 pKa = 3.37NALLHH44 pKa = 6.05FARR47 pKa = 11.84QKK49 pKa = 10.05GLSRR53 pKa = 11.84IDD55 pKa = 3.58LTPVPSLAASACKK68 pKa = 9.89AKK70 pKa = 10.31QAIEE74 pKa = 4.2IEE76 pKa = 4.24LSLEE80 pKa = 4.39GIKK83 pKa = 10.84NSGWDD88 pKa = 3.71DD89 pKa = 4.21EE90 pKa = 4.67PWTLQDD96 pKa = 4.35GSWDD100 pKa = 3.58RR101 pKa = 11.84YY102 pKa = 9.01AASPEE107 pKa = 4.03GAFKK111 pKa = 10.94KK112 pKa = 10.55NPAVVEE118 pKa = 4.55VIFDD122 pKa = 4.08EE123 pKa = 4.47NPEE126 pKa = 3.87NAVWYY131 pKa = 6.2THH133 pKa = 6.42WEE135 pKa = 3.98VLYY138 pKa = 9.9VTDD141 pKa = 3.94GVKK144 pKa = 8.75WHH146 pKa = 6.55RR147 pKa = 11.84TTGRR151 pKa = 11.84VCANGLFYY159 pKa = 10.78EE160 pKa = 4.77LGGEE164 pKa = 3.88QTFYY168 pKa = 11.52EE169 pKa = 4.72RR170 pKa = 11.84FDD172 pKa = 4.54LDD174 pKa = 3.37AQKK177 pKa = 11.16YY178 pKa = 6.09STGGTWTVTFQGEE191 pKa = 4.66TMTGSLVSSSTDD203 pKa = 3.25GVDD206 pKa = 3.59GEE208 pKa = 4.79ARR210 pKa = 11.84PPSPMPASTACGEE223 pKa = 4.1RR224 pKa = 11.84RR225 pKa = 11.84DD226 pKa = 4.15NLSTPAEE233 pKa = 4.41PDD235 pKa = 2.94SSSAVSSNFTSSGGRR250 pKa = 11.84SRR252 pKa = 11.84GRR254 pKa = 11.84ANRR257 pKa = 11.84GCPYY261 pKa = 10.09TRR263 pKa = 11.84TEE265 pKa = 4.21GPRR268 pKa = 11.84VPKK271 pKa = 10.44VVSAPCYY278 pKa = 9.69RR279 pKa = 11.84PRR281 pKa = 11.84GVQAQLGSTPPLVSPADD298 pKa = 3.57PLGRR302 pKa = 11.84SSAGEE307 pKa = 4.17RR308 pKa = 11.84PPGAAGHH315 pKa = 6.17SGSVGNAVPPAAPPASPFLQGSRR338 pKa = 11.84CPVILVTGSCNQVKK352 pKa = 9.74CWRR355 pKa = 11.84LRR357 pKa = 11.84CRR359 pKa = 11.84RR360 pKa = 11.84HH361 pKa = 5.06HH362 pKa = 5.27RR363 pKa = 11.84HH364 pKa = 5.93RR365 pKa = 11.84YY366 pKa = 8.44HH367 pKa = 7.2RR368 pKa = 11.84VTSTWYY374 pKa = 8.43ITGDD378 pKa = 3.38QGNDD382 pKa = 2.83RR383 pKa = 11.84AGPSTVMVMFTDD395 pKa = 4.24DD396 pKa = 3.26AQRR399 pKa = 11.84ADD401 pKa = 4.05FMAKK405 pKa = 9.6VSFPPGVSCTPLTLSTT421 pKa = 4.27
Molecular weight: 45.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2204 |
99 |
625 |
440.8 |
48.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.258 ± 0.379 | 1.906 ± 0.685 |
5.944 ± 0.255 | 6.488 ± 0.39 |
4.083 ± 0.456 | 7.123 ± 0.705 |
2.269 ± 0.303 | 3.539 ± 0.441 |
3.857 ± 0.662 | 8.848 ± 0.556 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.497 ± 0.137 | 3.993 ± 0.631 |
6.987 ± 0.708 | 3.766 ± 0.315 |
6.67 ± 0.246 | 7.577 ± 0.706 |
6.216 ± 0.437 | 6.261 ± 0.614 |
1.679 ± 0.306 | 3.04 ± 0.273 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
