
Croceicoccus naphthovorans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Croceicoccus
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
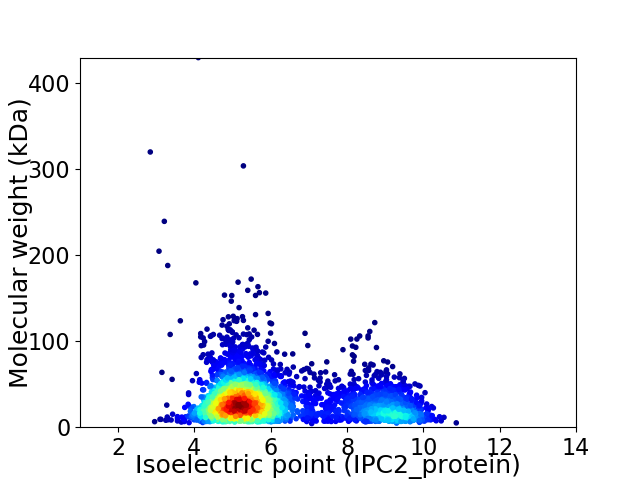
Virtual 2D-PAGE plot for 3411 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
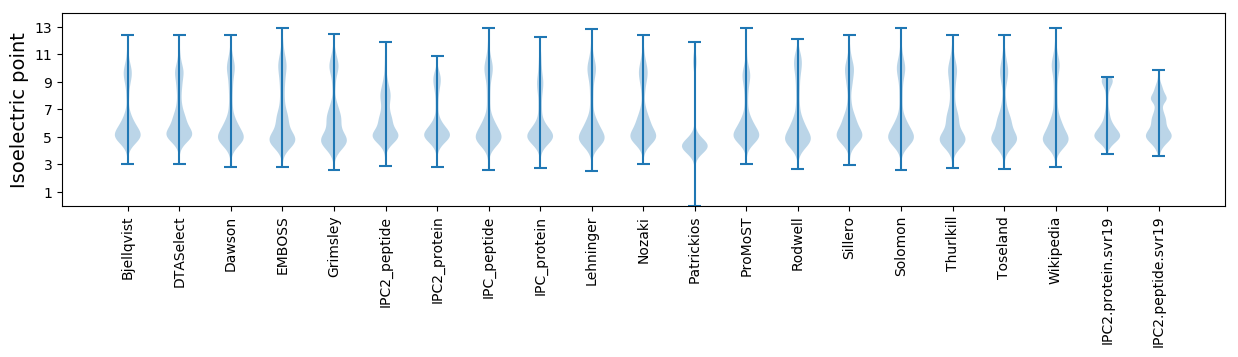
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G3XKA2|A0A0G3XKA2_9SPHN Conjugal transfer pilus assembly protein TraB OS=Croceicoccus naphthovorans OX=1348774 GN=AB433_17790 PE=4 SV=1
MM1 pKa = 7.49LSAACSQAVADD12 pKa = 4.56EE13 pKa = 4.5PMPADD18 pKa = 5.05EE19 pKa = 4.93IAAPDD24 pKa = 3.89AQGMAEE30 pKa = 4.79DD31 pKa = 4.96GDD33 pKa = 4.51PNGAACEE40 pKa = 3.98AAGGFLDD47 pKa = 4.27RR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AQTLMCVHH59 pKa = 7.52PYY61 pKa = 11.01ADD63 pKa = 3.94AGKK66 pKa = 10.31ACTDD70 pKa = 3.32NQQCDD75 pKa = 4.48GKK77 pKa = 10.87CVAAVDD83 pKa = 4.57DD84 pKa = 5.0GPDD87 pKa = 3.62GEE89 pKa = 5.44VVGTCQADD97 pKa = 3.45DD98 pKa = 4.36ALFGCYY104 pKa = 10.13AEE106 pKa = 4.75VVDD109 pKa = 4.78GKK111 pKa = 9.98LVRR114 pKa = 11.84AICVDD119 pKa = 3.34
MM1 pKa = 7.49LSAACSQAVADD12 pKa = 4.56EE13 pKa = 4.5PMPADD18 pKa = 5.05EE19 pKa = 4.93IAAPDD24 pKa = 3.89AQGMAEE30 pKa = 4.79DD31 pKa = 4.96GDD33 pKa = 4.51PNGAACEE40 pKa = 3.98AAGGFLDD47 pKa = 4.27RR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AQTLMCVHH59 pKa = 7.52PYY61 pKa = 11.01ADD63 pKa = 3.94AGKK66 pKa = 10.31ACTDD70 pKa = 3.32NQQCDD75 pKa = 4.48GKK77 pKa = 10.87CVAAVDD83 pKa = 4.57DD84 pKa = 5.0GPDD87 pKa = 3.62GEE89 pKa = 5.44VVGTCQADD97 pKa = 3.45DD98 pKa = 4.36ALFGCYY104 pKa = 10.13AEE106 pKa = 4.75VVDD109 pKa = 4.78GKK111 pKa = 9.98LVRR114 pKa = 11.84AICVDD119 pKa = 3.34
Molecular weight: 12.07 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G3XNL2|A0A0G3XNL2_9SPHN Conjugal transfer pilus assembly protein TraV OS=Croceicoccus naphthovorans OX=1348774 GN=AB433_18890 PE=4 SV=1
MM1 pKa = 7.43TLACNRR7 pKa = 11.84PTSEE11 pKa = 4.08LKK13 pKa = 10.97ALVSRR18 pKa = 11.84LGGTWSGNTAMCLCPAHH35 pKa = 7.3ADD37 pKa = 3.87RR38 pKa = 11.84TPSLSIRR45 pKa = 11.84QGDD48 pKa = 3.71RR49 pKa = 11.84AILVTCHH56 pKa = 6.66AGCDD60 pKa = 3.39RR61 pKa = 11.84SDD63 pKa = 3.31VLRR66 pKa = 11.84AIGRR70 pKa = 11.84ITRR73 pKa = 11.84IPHH76 pKa = 6.12FDD78 pKa = 3.25PAKK81 pKa = 10.24IEE83 pKa = 3.98RR84 pKa = 11.84APARR88 pKa = 11.84SRR90 pKa = 11.84NAFLKK95 pKa = 9.5IWRR98 pKa = 11.84EE99 pKa = 3.61GRR101 pKa = 11.84PIEE104 pKa = 4.4GSLAEE109 pKa = 4.21YY110 pKa = 10.11YY111 pKa = 10.24VRR113 pKa = 11.84QVRR116 pKa = 11.84GIGGVLQDD124 pKa = 4.13LRR126 pKa = 11.84FHH128 pKa = 6.63PRR130 pKa = 11.84CPRR133 pKa = 11.84GQGALARR140 pKa = 11.84FEE142 pKa = 4.19PALLVGMRR150 pKa = 11.84RR151 pKa = 11.84DD152 pKa = 3.39GNLAAIQRR160 pKa = 11.84IFLDD164 pKa = 3.66PRR166 pKa = 11.84TGASTAKK173 pKa = 10.07LCLGRR178 pKa = 11.84AIGAAWTNGTPEE190 pKa = 4.28SVLGLCEE197 pKa = 4.08GFEE200 pKa = 4.13TAAAFTDD207 pKa = 3.99LVGIKK212 pKa = 9.63AWASMGAKK220 pKa = 9.72RR221 pKa = 11.84FHH223 pKa = 6.45QLTIPRR229 pKa = 11.84TVVRR233 pKa = 11.84LILLADD239 pKa = 3.82NDD241 pKa = 4.56AEE243 pKa = 4.32GHH245 pKa = 5.65RR246 pKa = 11.84AANRR250 pKa = 11.84ALAAYY255 pKa = 9.2SRR257 pKa = 11.84SGLAIEE263 pKa = 4.7TRR265 pKa = 11.84WPPRR269 pKa = 11.84GANDD273 pKa = 3.29WADD276 pKa = 3.49LLKK279 pKa = 10.76RR280 pKa = 4.01
MM1 pKa = 7.43TLACNRR7 pKa = 11.84PTSEE11 pKa = 4.08LKK13 pKa = 10.97ALVSRR18 pKa = 11.84LGGTWSGNTAMCLCPAHH35 pKa = 7.3ADD37 pKa = 3.87RR38 pKa = 11.84TPSLSIRR45 pKa = 11.84QGDD48 pKa = 3.71RR49 pKa = 11.84AILVTCHH56 pKa = 6.66AGCDD60 pKa = 3.39RR61 pKa = 11.84SDD63 pKa = 3.31VLRR66 pKa = 11.84AIGRR70 pKa = 11.84ITRR73 pKa = 11.84IPHH76 pKa = 6.12FDD78 pKa = 3.25PAKK81 pKa = 10.24IEE83 pKa = 3.98RR84 pKa = 11.84APARR88 pKa = 11.84SRR90 pKa = 11.84NAFLKK95 pKa = 9.5IWRR98 pKa = 11.84EE99 pKa = 3.61GRR101 pKa = 11.84PIEE104 pKa = 4.4GSLAEE109 pKa = 4.21YY110 pKa = 10.11YY111 pKa = 10.24VRR113 pKa = 11.84QVRR116 pKa = 11.84GIGGVLQDD124 pKa = 4.13LRR126 pKa = 11.84FHH128 pKa = 6.63PRR130 pKa = 11.84CPRR133 pKa = 11.84GQGALARR140 pKa = 11.84FEE142 pKa = 4.19PALLVGMRR150 pKa = 11.84RR151 pKa = 11.84DD152 pKa = 3.39GNLAAIQRR160 pKa = 11.84IFLDD164 pKa = 3.66PRR166 pKa = 11.84TGASTAKK173 pKa = 10.07LCLGRR178 pKa = 11.84AIGAAWTNGTPEE190 pKa = 4.28SVLGLCEE197 pKa = 4.08GFEE200 pKa = 4.13TAAAFTDD207 pKa = 3.99LVGIKK212 pKa = 9.63AWASMGAKK220 pKa = 9.72RR221 pKa = 11.84FHH223 pKa = 6.45QLTIPRR229 pKa = 11.84TVVRR233 pKa = 11.84LILLADD239 pKa = 3.82NDD241 pKa = 4.56AEE243 pKa = 4.32GHH245 pKa = 5.65RR246 pKa = 11.84AANRR250 pKa = 11.84ALAAYY255 pKa = 9.2SRR257 pKa = 11.84SGLAIEE263 pKa = 4.7TRR265 pKa = 11.84WPPRR269 pKa = 11.84GANDD273 pKa = 3.29WADD276 pKa = 3.49LLKK279 pKa = 10.76RR280 pKa = 4.01
Molecular weight: 30.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1042385 |
40 |
4131 |
305.6 |
33.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.677 ± 0.061 | 0.845 ± 0.015 |
6.352 ± 0.039 | 5.984 ± 0.043 |
3.577 ± 0.025 | 8.773 ± 0.056 |
1.975 ± 0.022 | 5.087 ± 0.031 |
3.196 ± 0.041 | 9.642 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.56 ± 0.023 | 2.631 ± 0.027 |
5.083 ± 0.034 | 3.078 ± 0.022 |
7.134 ± 0.048 | 5.315 ± 0.035 |
5.28 ± 0.042 | 7.171 ± 0.033 |
1.427 ± 0.018 | 2.212 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
