
Shuangao insect virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
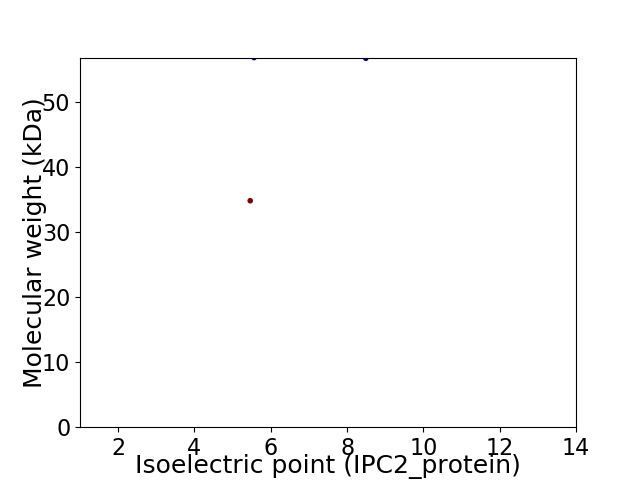
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEF4|A0A1L3KEF4_9VIRU RNA-directed RNA polymerase OS=Shuangao insect virus 9 OX=1923470 PE=4 SV=1
MM1 pKa = 7.93RR2 pKa = 11.84MLALEE7 pKa = 4.89AGDD10 pKa = 4.06LQADD14 pKa = 4.72HH15 pKa = 6.9IRR17 pKa = 11.84VFVKK21 pKa = 10.23PEE23 pKa = 3.5PHH25 pKa = 6.06KK26 pKa = 10.16QAKK29 pKa = 10.03LEE31 pKa = 3.99EE32 pKa = 4.39GRR34 pKa = 11.84LRR36 pKa = 11.84LISAVSLVDD45 pKa = 3.27TCVDD49 pKa = 4.23RR50 pKa = 11.84ILFTWCLEE58 pKa = 3.93AALDD62 pKa = 4.03NFLTTPSMIGWTPIYY77 pKa = 10.44GSWRR81 pKa = 11.84QLWALYY87 pKa = 10.31GGSALALDD95 pKa = 4.22KK96 pKa = 11.25SAWDD100 pKa = 3.33WTVQPWLIDD109 pKa = 4.27AIRR112 pKa = 11.84DD113 pKa = 3.9VIKK116 pKa = 10.45EE117 pKa = 3.98LAIDD121 pKa = 4.81APPWWQALVDD131 pKa = 4.04RR132 pKa = 11.84RR133 pKa = 11.84FEE135 pKa = 4.56LLFQSPVFEE144 pKa = 4.52FQDD147 pKa = 3.59GTTVKK152 pKa = 10.67QMGKK156 pKa = 10.83GIMKK160 pKa = 9.81SGCYY164 pKa = 8.43LTIIANTLGQVITHH178 pKa = 6.52FVACQQTGDD187 pKa = 4.0NPVLSVPHH195 pKa = 6.76ALGDD199 pKa = 3.74DD200 pKa = 4.44TIQRR204 pKa = 11.84GDD206 pKa = 3.86RR207 pKa = 11.84INDD210 pKa = 2.99IGLYY214 pKa = 9.75CAALEE219 pKa = 4.17QLGCKK224 pKa = 9.78VKK226 pKa = 10.56GGQVATDD233 pKa = 3.32VVEE236 pKa = 4.42FVGFEE241 pKa = 4.02FRR243 pKa = 11.84EE244 pKa = 4.48HH245 pKa = 6.11SCVPAYY251 pKa = 10.21RR252 pKa = 11.84EE253 pKa = 3.59KK254 pKa = 10.87HH255 pKa = 5.45FYY257 pKa = 9.51KK258 pKa = 10.62QEE260 pKa = 3.85FTPDD264 pKa = 3.07LGGFLYY270 pKa = 10.81AMQLLYY276 pKa = 11.14AHH278 pKa = 7.39DD279 pKa = 4.04PAFYY283 pKa = 10.14AYY285 pKa = 9.92YY286 pKa = 10.29RR287 pKa = 11.84RR288 pKa = 11.84LAAEE292 pKa = 4.23KK293 pKa = 9.84VPSRR297 pKa = 11.84VTSAIEE303 pKa = 3.85AKK305 pKa = 10.78GFMAA309 pKa = 6.13
MM1 pKa = 7.93RR2 pKa = 11.84MLALEE7 pKa = 4.89AGDD10 pKa = 4.06LQADD14 pKa = 4.72HH15 pKa = 6.9IRR17 pKa = 11.84VFVKK21 pKa = 10.23PEE23 pKa = 3.5PHH25 pKa = 6.06KK26 pKa = 10.16QAKK29 pKa = 10.03LEE31 pKa = 3.99EE32 pKa = 4.39GRR34 pKa = 11.84LRR36 pKa = 11.84LISAVSLVDD45 pKa = 3.27TCVDD49 pKa = 4.23RR50 pKa = 11.84ILFTWCLEE58 pKa = 3.93AALDD62 pKa = 4.03NFLTTPSMIGWTPIYY77 pKa = 10.44GSWRR81 pKa = 11.84QLWALYY87 pKa = 10.31GGSALALDD95 pKa = 4.22KK96 pKa = 11.25SAWDD100 pKa = 3.33WTVQPWLIDD109 pKa = 4.27AIRR112 pKa = 11.84DD113 pKa = 3.9VIKK116 pKa = 10.45EE117 pKa = 3.98LAIDD121 pKa = 4.81APPWWQALVDD131 pKa = 4.04RR132 pKa = 11.84RR133 pKa = 11.84FEE135 pKa = 4.56LLFQSPVFEE144 pKa = 4.52FQDD147 pKa = 3.59GTTVKK152 pKa = 10.67QMGKK156 pKa = 10.83GIMKK160 pKa = 9.81SGCYY164 pKa = 8.43LTIIANTLGQVITHH178 pKa = 6.52FVACQQTGDD187 pKa = 4.0NPVLSVPHH195 pKa = 6.76ALGDD199 pKa = 3.74DD200 pKa = 4.44TIQRR204 pKa = 11.84GDD206 pKa = 3.86RR207 pKa = 11.84INDD210 pKa = 2.99IGLYY214 pKa = 9.75CAALEE219 pKa = 4.17QLGCKK224 pKa = 9.78VKK226 pKa = 10.56GGQVATDD233 pKa = 3.32VVEE236 pKa = 4.42FVGFEE241 pKa = 4.02FRR243 pKa = 11.84EE244 pKa = 4.48HH245 pKa = 6.11SCVPAYY251 pKa = 10.21RR252 pKa = 11.84EE253 pKa = 3.59KK254 pKa = 10.87HH255 pKa = 5.45FYY257 pKa = 9.51KK258 pKa = 10.62QEE260 pKa = 3.85FTPDD264 pKa = 3.07LGGFLYY270 pKa = 10.81AMQLLYY276 pKa = 11.14AHH278 pKa = 7.39DD279 pKa = 4.04PAFYY283 pKa = 10.14AYY285 pKa = 9.92YY286 pKa = 10.29RR287 pKa = 11.84RR288 pKa = 11.84LAAEE292 pKa = 4.23KK293 pKa = 9.84VPSRR297 pKa = 11.84VTSAIEE303 pKa = 3.85AKK305 pKa = 10.78GFMAA309 pKa = 6.13
Molecular weight: 34.82 kDa
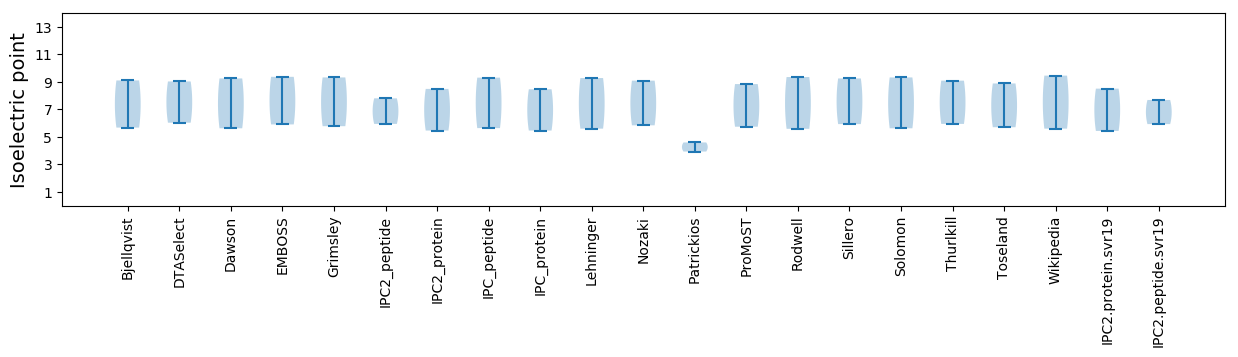
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEF4|A0A1L3KEF4_9VIRU RNA-directed RNA polymerase OS=Shuangao insect virus 9 OX=1923470 PE=4 SV=1
MM1 pKa = 7.6LWFSSQPDD9 pKa = 3.25KK10 pKa = 10.94MVTVSFEE17 pKa = 3.9IPEE20 pKa = 4.23KK21 pKa = 10.53KK22 pKa = 9.73VSKK25 pKa = 10.4VKK27 pKa = 10.66LVTRR31 pKa = 11.84YY32 pKa = 8.45GWGWYY37 pKa = 9.84RR38 pKa = 11.84FIGLFWPIIVASQLYY53 pKa = 8.85MLAFYY58 pKa = 10.9LSLDD62 pKa = 3.09ATTWQFAFSQTVAEE76 pKa = 4.1PVVRR80 pKa = 11.84LVNAVFSTLVLCFVVYY96 pKa = 10.2LMRR99 pKa = 11.84GTMRR103 pKa = 11.84LTGLCKK109 pKa = 10.61DD110 pKa = 3.61PTGSVTFEE118 pKa = 4.25PEE120 pKa = 3.32RR121 pKa = 11.84MRR123 pKa = 11.84PGSLLMPHH131 pKa = 7.13APSKK135 pKa = 10.68CQAEE139 pKa = 4.81LYY141 pKa = 10.79GKK143 pKa = 7.29TQGTWVRR150 pKa = 11.84LGQAFRR156 pKa = 11.84SSNYY160 pKa = 10.07LVTASHH166 pKa = 7.07VIDD169 pKa = 3.33QVEE172 pKa = 4.17IVKK175 pKa = 8.67ITTEE179 pKa = 3.79HH180 pKa = 6.71ASIEE184 pKa = 4.45VPTSLFRR191 pKa = 11.84QVEE194 pKa = 4.29GDD196 pKa = 3.76LSIMPVTDD204 pKa = 3.53QVLAPLKK211 pKa = 10.12LANGRR216 pKa = 11.84LASHH220 pKa = 7.05EE221 pKa = 4.06ATEE224 pKa = 4.55GSGLFVKK231 pKa = 10.53IGGFGSGSLGMLKK244 pKa = 10.2PNPAFGFVEE253 pKa = 4.55YY254 pKa = 9.97TGSTVPGFSGAPYY267 pKa = 10.14VVGDD271 pKa = 4.1VIYY274 pKa = 10.59GMHH277 pKa = 7.04LGGGAQNLGYY287 pKa = 9.9SGAYY291 pKa = 9.27ISMLMRR297 pKa = 11.84ALQEE301 pKa = 4.44DD302 pKa = 3.88STDD305 pKa = 3.65FLMSQIEE312 pKa = 4.08RR313 pKa = 11.84AAEE316 pKa = 4.01YY317 pKa = 10.15QVSQSPYY324 pKa = 9.85DD325 pKa = 3.76PDD327 pKa = 3.36EE328 pKa = 4.52FRR330 pKa = 11.84VKK332 pKa = 10.78VGGRR336 pKa = 11.84YY337 pKa = 9.43YY338 pKa = 11.12LLDD341 pKa = 3.89AEE343 pKa = 5.31GVHH346 pKa = 6.41KK347 pKa = 11.02LNLRR351 pKa = 11.84VQRR354 pKa = 11.84DD355 pKa = 3.53VSSGYY360 pKa = 10.05RR361 pKa = 11.84DD362 pKa = 3.86YY363 pKa = 11.2EE364 pKa = 4.14PEE366 pKa = 4.08GALPQVEE373 pKa = 4.53KK374 pKa = 10.82VVKK377 pKa = 9.86PVVIPATSPKK387 pKa = 10.03PPMARR392 pKa = 11.84VEE394 pKa = 4.18EE395 pKa = 4.58LEE397 pKa = 4.83SLLQTGNGPAPAEE410 pKa = 4.17NPVPAGATGQDD421 pKa = 3.57SVPARR426 pKa = 11.84PARR429 pKa = 11.84RR430 pKa = 11.84PRR432 pKa = 11.84RR433 pKa = 11.84RR434 pKa = 11.84SPAATTSSATLPSSATDD451 pKa = 3.96GQDD454 pKa = 2.95QTPAQLRR461 pKa = 11.84GALQDD466 pKa = 3.37IADD469 pKa = 4.16SLKK472 pKa = 10.02MLRR475 pKa = 11.84KK476 pKa = 9.48EE477 pKa = 4.1LGTLQMQTSRR487 pKa = 11.84RR488 pKa = 11.84RR489 pKa = 11.84LKK491 pKa = 10.4EE492 pKa = 3.98SEE494 pKa = 4.26SASATGRR501 pKa = 11.84LTSLRR506 pKa = 11.84RR507 pKa = 11.84EE508 pKa = 4.23LSRR511 pKa = 11.84LRR513 pKa = 11.84SSGGSQQ519 pKa = 2.98
MM1 pKa = 7.6LWFSSQPDD9 pKa = 3.25KK10 pKa = 10.94MVTVSFEE17 pKa = 3.9IPEE20 pKa = 4.23KK21 pKa = 10.53KK22 pKa = 9.73VSKK25 pKa = 10.4VKK27 pKa = 10.66LVTRR31 pKa = 11.84YY32 pKa = 8.45GWGWYY37 pKa = 9.84RR38 pKa = 11.84FIGLFWPIIVASQLYY53 pKa = 8.85MLAFYY58 pKa = 10.9LSLDD62 pKa = 3.09ATTWQFAFSQTVAEE76 pKa = 4.1PVVRR80 pKa = 11.84LVNAVFSTLVLCFVVYY96 pKa = 10.2LMRR99 pKa = 11.84GTMRR103 pKa = 11.84LTGLCKK109 pKa = 10.61DD110 pKa = 3.61PTGSVTFEE118 pKa = 4.25PEE120 pKa = 3.32RR121 pKa = 11.84MRR123 pKa = 11.84PGSLLMPHH131 pKa = 7.13APSKK135 pKa = 10.68CQAEE139 pKa = 4.81LYY141 pKa = 10.79GKK143 pKa = 7.29TQGTWVRR150 pKa = 11.84LGQAFRR156 pKa = 11.84SSNYY160 pKa = 10.07LVTASHH166 pKa = 7.07VIDD169 pKa = 3.33QVEE172 pKa = 4.17IVKK175 pKa = 8.67ITTEE179 pKa = 3.79HH180 pKa = 6.71ASIEE184 pKa = 4.45VPTSLFRR191 pKa = 11.84QVEE194 pKa = 4.29GDD196 pKa = 3.76LSIMPVTDD204 pKa = 3.53QVLAPLKK211 pKa = 10.12LANGRR216 pKa = 11.84LASHH220 pKa = 7.05EE221 pKa = 4.06ATEE224 pKa = 4.55GSGLFVKK231 pKa = 10.53IGGFGSGSLGMLKK244 pKa = 10.2PNPAFGFVEE253 pKa = 4.55YY254 pKa = 9.97TGSTVPGFSGAPYY267 pKa = 10.14VVGDD271 pKa = 4.1VIYY274 pKa = 10.59GMHH277 pKa = 7.04LGGGAQNLGYY287 pKa = 9.9SGAYY291 pKa = 9.27ISMLMRR297 pKa = 11.84ALQEE301 pKa = 4.44DD302 pKa = 3.88STDD305 pKa = 3.65FLMSQIEE312 pKa = 4.08RR313 pKa = 11.84AAEE316 pKa = 4.01YY317 pKa = 10.15QVSQSPYY324 pKa = 9.85DD325 pKa = 3.76PDD327 pKa = 3.36EE328 pKa = 4.52FRR330 pKa = 11.84VKK332 pKa = 10.78VGGRR336 pKa = 11.84YY337 pKa = 9.43YY338 pKa = 11.12LLDD341 pKa = 3.89AEE343 pKa = 5.31GVHH346 pKa = 6.41KK347 pKa = 11.02LNLRR351 pKa = 11.84VQRR354 pKa = 11.84DD355 pKa = 3.53VSSGYY360 pKa = 10.05RR361 pKa = 11.84DD362 pKa = 3.86YY363 pKa = 11.2EE364 pKa = 4.14PEE366 pKa = 4.08GALPQVEE373 pKa = 4.53KK374 pKa = 10.82VVKK377 pKa = 9.86PVVIPATSPKK387 pKa = 10.03PPMARR392 pKa = 11.84VEE394 pKa = 4.18EE395 pKa = 4.58LEE397 pKa = 4.83SLLQTGNGPAPAEE410 pKa = 4.17NPVPAGATGQDD421 pKa = 3.57SVPARR426 pKa = 11.84PARR429 pKa = 11.84RR430 pKa = 11.84PRR432 pKa = 11.84RR433 pKa = 11.84RR434 pKa = 11.84SPAATTSSATLPSSATDD451 pKa = 3.96GQDD454 pKa = 2.95QTPAQLRR461 pKa = 11.84GALQDD466 pKa = 3.37IADD469 pKa = 4.16SLKK472 pKa = 10.02MLRR475 pKa = 11.84KK476 pKa = 9.48EE477 pKa = 4.1LGTLQMQTSRR487 pKa = 11.84RR488 pKa = 11.84RR489 pKa = 11.84LKK491 pKa = 10.4EE492 pKa = 3.98SEE494 pKa = 4.26SASATGRR501 pKa = 11.84LTSLRR506 pKa = 11.84RR507 pKa = 11.84EE508 pKa = 4.23LSRR511 pKa = 11.84LRR513 pKa = 11.84SSGGSQQ519 pKa = 2.98
Molecular weight: 56.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
828 |
309 |
519 |
414.0 |
45.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.696 ± 0.789 | 1.208 ± 0.624 |
4.71 ± 1.04 | 5.435 ± 0.152 |
4.227 ± 0.561 | 8.092 ± 0.574 |
1.57 ± 0.41 | 3.865 ± 0.966 |
4.106 ± 0.25 | 10.145 ± 0.125 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.778 ± 0.302 | 1.449 ± 0.091 |
5.918 ± 0.818 | 5.072 ± 0.062 |
6.159 ± 0.579 | 7.367 ± 2.055 |
5.918 ± 0.437 | 7.971 ± 0.502 |
1.812 ± 0.65 | 3.502 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
