
Faeces associated gemycircularvirus 19
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus porci1; Porcine associated gemycircularvirus 1
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
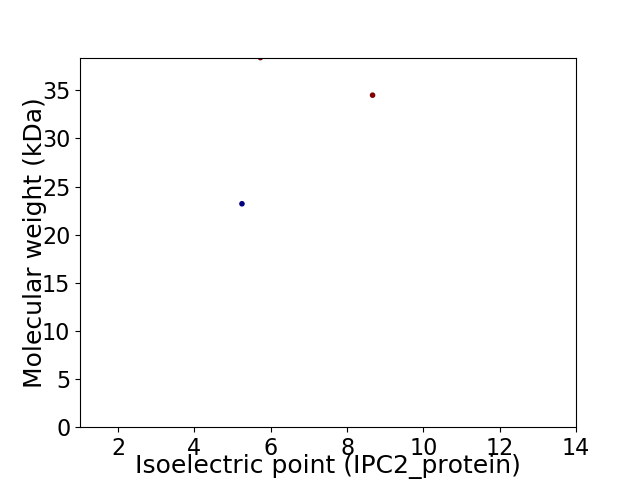
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
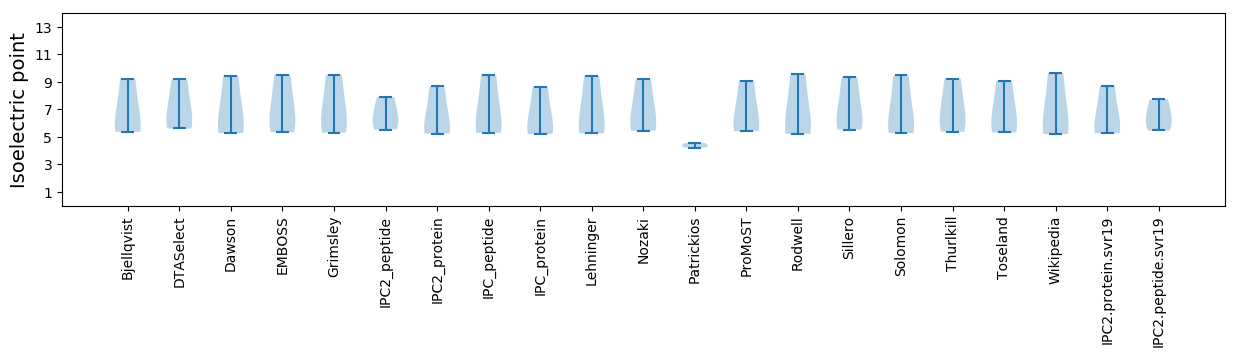
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A168MGA2|A0A168MGA2_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 19 OX=1843739 PE=3 SV=1
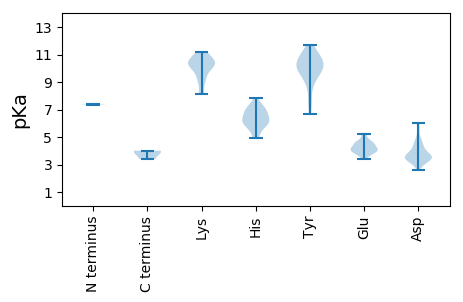
MM1 pKa = 7.4SAFHH5 pKa = 6.68FSARR9 pKa = 11.84YY10 pKa = 9.45VLLTYY15 pKa = 9.0PQSGEE20 pKa = 3.86LSEE23 pKa = 4.35WAVLDD28 pKa = 4.71HH29 pKa = 7.1ISGLGAEE36 pKa = 4.74CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.04LHH52 pKa = 5.93VFADD56 pKa = 4.85FGRR59 pKa = 11.84KK60 pKa = 8.21KK61 pKa = 9.8QSRR64 pKa = 11.84RR65 pKa = 11.84GDD67 pKa = 3.58YY68 pKa = 10.34FDD70 pKa = 5.37VGGKK74 pKa = 9.22HH75 pKa = 6.11PNVVPSKK82 pKa = 10.3GRR84 pKa = 11.84PEE86 pKa = 4.58GGWDD90 pKa = 3.41YY91 pKa = 10.93ATKK94 pKa = 10.52DD95 pKa = 3.6GNVVAGGLGRR105 pKa = 11.84PGTSGLPKK113 pKa = 10.41APNPWRR119 pKa = 11.84EE120 pKa = 3.71IVGAEE125 pKa = 3.81GRR127 pKa = 11.84EE128 pKa = 3.91EE129 pKa = 4.09FLDD132 pKa = 4.45LVRR135 pKa = 11.84QLDD138 pKa = 3.77PKK140 pKa = 11.22SFVLKK145 pKa = 9.09HH146 pKa = 4.95QEE148 pKa = 3.38IVRR151 pKa = 11.84YY152 pKa = 9.92ADD154 pKa = 2.87IFFAEE159 pKa = 4.26DD160 pKa = 3.23RR161 pKa = 11.84EE162 pKa = 4.59PYY164 pKa = 10.05VGPDD168 pKa = 4.0GIRR171 pKa = 11.84FEE173 pKa = 5.22LGMVPQLDD181 pKa = 3.24EE182 pKa = 3.93WRR184 pKa = 11.84RR185 pKa = 11.84EE186 pKa = 4.08SLGDD190 pKa = 3.43NPVEE194 pKa = 4.57GKK196 pKa = 8.11CTPSLASSGGPGTDD210 pKa = 3.53LSS212 pKa = 4.01
MM1 pKa = 7.4SAFHH5 pKa = 6.68FSARR9 pKa = 11.84YY10 pKa = 9.45VLLTYY15 pKa = 9.0PQSGEE20 pKa = 3.86LSEE23 pKa = 4.35WAVLDD28 pKa = 4.71HH29 pKa = 7.1ISGLGAEE36 pKa = 4.74CIIGRR41 pKa = 11.84EE42 pKa = 4.05DD43 pKa = 3.35HH44 pKa = 7.18ADD46 pKa = 3.64GGTHH50 pKa = 6.04LHH52 pKa = 5.93VFADD56 pKa = 4.85FGRR59 pKa = 11.84KK60 pKa = 8.21KK61 pKa = 9.8QSRR64 pKa = 11.84RR65 pKa = 11.84GDD67 pKa = 3.58YY68 pKa = 10.34FDD70 pKa = 5.37VGGKK74 pKa = 9.22HH75 pKa = 6.11PNVVPSKK82 pKa = 10.3GRR84 pKa = 11.84PEE86 pKa = 4.58GGWDD90 pKa = 3.41YY91 pKa = 10.93ATKK94 pKa = 10.52DD95 pKa = 3.6GNVVAGGLGRR105 pKa = 11.84PGTSGLPKK113 pKa = 10.41APNPWRR119 pKa = 11.84EE120 pKa = 3.71IVGAEE125 pKa = 3.81GRR127 pKa = 11.84EE128 pKa = 3.91EE129 pKa = 4.09FLDD132 pKa = 4.45LVRR135 pKa = 11.84QLDD138 pKa = 3.77PKK140 pKa = 11.22SFVLKK145 pKa = 9.09HH146 pKa = 4.95QEE148 pKa = 3.38IVRR151 pKa = 11.84YY152 pKa = 9.92ADD154 pKa = 2.87IFFAEE159 pKa = 4.26DD160 pKa = 3.23RR161 pKa = 11.84EE162 pKa = 4.59PYY164 pKa = 10.05VGPDD168 pKa = 4.0GIRR171 pKa = 11.84FEE173 pKa = 5.22LGMVPQLDD181 pKa = 3.24EE182 pKa = 3.93WRR184 pKa = 11.84RR185 pKa = 11.84EE186 pKa = 4.08SLGDD190 pKa = 3.43NPVEE194 pKa = 4.57GKK196 pKa = 8.11CTPSLASSGGPGTDD210 pKa = 3.53LSS212 pKa = 4.01
Molecular weight: 23.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MG89|A0A168MG89_9VIRU RepA OS=Faeces associated gemycircularvirus 19 OX=1843739 PE=3 SV=1
MM1 pKa = 7.36PRR3 pKa = 11.84YY4 pKa = 9.42RR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 9.81RR8 pKa = 11.84AQPRR12 pKa = 11.84LKK14 pKa = 10.36RR15 pKa = 11.84RR16 pKa = 11.84MKK18 pKa = 10.45KK19 pKa = 10.22AVLNTTSTKK28 pKa = 10.35KK29 pKa = 9.95RR30 pKa = 11.84DD31 pKa = 3.47NMTPISNTTALGAPTPNILRR51 pKa = 11.84NGAVVIKK58 pKa = 10.78GGDD61 pKa = 3.51PSSAAGAGVSGAIFVWSPTARR82 pKa = 11.84PMNDD86 pKa = 2.57NTNAEE91 pKa = 4.16NTRR94 pKa = 11.84VKK96 pKa = 10.5RR97 pKa = 11.84DD98 pKa = 3.36VYY100 pKa = 11.06HH101 pKa = 6.45KK102 pKa = 10.78GFRR105 pKa = 11.84EE106 pKa = 4.02KK107 pKa = 11.12VKK109 pKa = 10.76VEE111 pKa = 4.49SNSSHH116 pKa = 5.57PWIWRR121 pKa = 11.84RR122 pKa = 11.84IVVEE126 pKa = 4.15TKK128 pKa = 10.27TDD130 pKa = 3.36DD131 pKa = 3.51FNYY134 pKa = 10.37LGEE137 pKa = 5.02SEE139 pKa = 5.08DD140 pKa = 4.32QPTLPPRR147 pKa = 11.84AFQPYY152 pKa = 7.77YY153 pKa = 10.27QGGEE157 pKa = 4.23GVSRR161 pKa = 11.84LWYY164 pKa = 8.64NHH166 pKa = 5.64YY167 pKa = 11.06GNRR170 pKa = 11.84TGPTDD175 pKa = 3.4TRR177 pKa = 11.84IDD179 pKa = 3.79SVLKK183 pKa = 10.0TIADD187 pKa = 3.82DD188 pKa = 4.62LFEE191 pKa = 4.48GTQNHH196 pKa = 6.73DD197 pKa = 3.19WLNTVTAKK205 pKa = 10.5VDD207 pKa = 3.62LDD209 pKa = 4.23RR210 pKa = 11.84YY211 pKa = 6.69TLRR214 pKa = 11.84YY215 pKa = 10.19DD216 pKa = 3.0KK217 pKa = 11.09TRR219 pKa = 11.84VFRR222 pKa = 11.84SGNDD226 pKa = 2.85AGFIKK231 pKa = 9.91TVNFYY236 pKa = 11.08HH237 pKa = 7.05SFEE240 pKa = 3.73KK241 pKa = 9.92TMYY244 pKa = 9.72YY245 pKa = 10.8DD246 pKa = 3.57HH247 pKa = 7.83DD248 pKa = 4.09EE249 pKa = 4.68DD250 pKa = 5.88RR251 pKa = 11.84NGTNPQSWVCAPALKK266 pKa = 10.77GMGNVYY272 pKa = 10.15IIDD275 pKa = 4.26IIEE278 pKa = 4.29SGISADD284 pKa = 3.46DD285 pKa = 3.44TDD287 pKa = 4.1YY288 pKa = 11.72LRR290 pKa = 11.84ITPEE294 pKa = 3.56SCAYY298 pKa = 7.48WHH300 pKa = 6.44EE301 pKa = 4.3RR302 pKa = 3.41
MM1 pKa = 7.36PRR3 pKa = 11.84YY4 pKa = 9.42RR5 pKa = 11.84RR6 pKa = 11.84YY7 pKa = 9.81RR8 pKa = 11.84AQPRR12 pKa = 11.84LKK14 pKa = 10.36RR15 pKa = 11.84RR16 pKa = 11.84MKK18 pKa = 10.45KK19 pKa = 10.22AVLNTTSTKK28 pKa = 10.35KK29 pKa = 9.95RR30 pKa = 11.84DD31 pKa = 3.47NMTPISNTTALGAPTPNILRR51 pKa = 11.84NGAVVIKK58 pKa = 10.78GGDD61 pKa = 3.51PSSAAGAGVSGAIFVWSPTARR82 pKa = 11.84PMNDD86 pKa = 2.57NTNAEE91 pKa = 4.16NTRR94 pKa = 11.84VKK96 pKa = 10.5RR97 pKa = 11.84DD98 pKa = 3.36VYY100 pKa = 11.06HH101 pKa = 6.45KK102 pKa = 10.78GFRR105 pKa = 11.84EE106 pKa = 4.02KK107 pKa = 11.12VKK109 pKa = 10.76VEE111 pKa = 4.49SNSSHH116 pKa = 5.57PWIWRR121 pKa = 11.84RR122 pKa = 11.84IVVEE126 pKa = 4.15TKK128 pKa = 10.27TDD130 pKa = 3.36DD131 pKa = 3.51FNYY134 pKa = 10.37LGEE137 pKa = 5.02SEE139 pKa = 5.08DD140 pKa = 4.32QPTLPPRR147 pKa = 11.84AFQPYY152 pKa = 7.77YY153 pKa = 10.27QGGEE157 pKa = 4.23GVSRR161 pKa = 11.84LWYY164 pKa = 8.64NHH166 pKa = 5.64YY167 pKa = 11.06GNRR170 pKa = 11.84TGPTDD175 pKa = 3.4TRR177 pKa = 11.84IDD179 pKa = 3.79SVLKK183 pKa = 10.0TIADD187 pKa = 3.82DD188 pKa = 4.62LFEE191 pKa = 4.48GTQNHH196 pKa = 6.73DD197 pKa = 3.19WLNTVTAKK205 pKa = 10.5VDD207 pKa = 3.62LDD209 pKa = 4.23RR210 pKa = 11.84YY211 pKa = 6.69TLRR214 pKa = 11.84YY215 pKa = 10.19DD216 pKa = 3.0KK217 pKa = 11.09TRR219 pKa = 11.84VFRR222 pKa = 11.84SGNDD226 pKa = 2.85AGFIKK231 pKa = 9.91TVNFYY236 pKa = 11.08HH237 pKa = 7.05SFEE240 pKa = 3.73KK241 pKa = 9.92TMYY244 pKa = 9.72YY245 pKa = 10.8DD246 pKa = 3.57HH247 pKa = 7.83DD248 pKa = 4.09EE249 pKa = 4.68DD250 pKa = 5.88RR251 pKa = 11.84NGTNPQSWVCAPALKK266 pKa = 10.77GMGNVYY272 pKa = 10.15IIDD275 pKa = 4.26IIEE278 pKa = 4.29SGISADD284 pKa = 3.46DD285 pKa = 3.44TDD287 pKa = 4.1YY288 pKa = 11.72LRR290 pKa = 11.84ITPEE294 pKa = 3.56SCAYY298 pKa = 7.48WHH300 pKa = 6.44EE301 pKa = 4.3RR302 pKa = 3.41
Molecular weight: 34.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
857 |
212 |
343 |
285.7 |
32.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.301 ± 0.068 | 0.817 ± 0.068 |
7.468 ± 0.09 | 5.951 ± 0.75 |
4.084 ± 0.474 | 10.152 ± 1.521 |
2.684 ± 0.223 | 4.201 ± 0.374 |
4.901 ± 0.326 | 7.118 ± 1.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.867 ± 0.335 | 3.734 ± 1.242 |
6.534 ± 0.389 | 2.217 ± 0.099 |
7.818 ± 0.358 | 6.301 ± 0.152 |
4.784 ± 1.791 | 6.884 ± 0.261 |
2.334 ± 0.177 | 3.851 ± 0.625 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
