
Psathyrella aberdarensis
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Psathyrellaceae; Candolleomyces
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
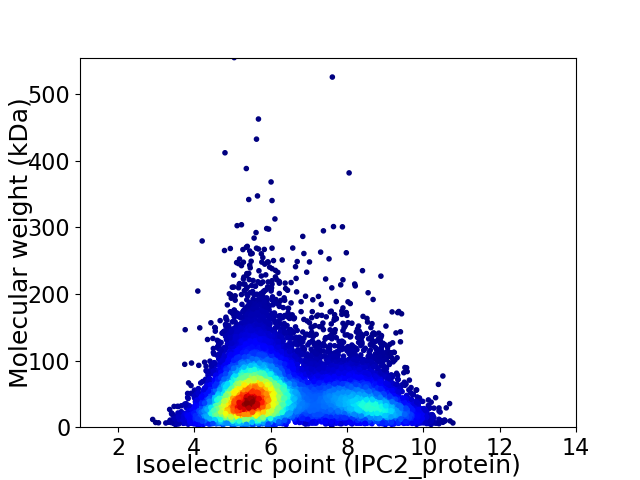
Virtual 2D-PAGE plot for 14686 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2D6S1|A0A4Q2D6S1_9AGAR GST N-terminal domain-containing protein OS=Psathyrella aberdarensis OX=2316362 GN=EST38_g10682 PE=4 SV=1
MM1 pKa = 7.51VKK3 pKa = 9.59TNPFNFVIAILFASFLHH20 pKa = 6.56VSAQTYY26 pKa = 7.66TVTNRR31 pKa = 11.84CPSAIEE37 pKa = 4.06LFIGQTSEE45 pKa = 4.69GNLATNQSIVKK56 pKa = 8.8TGLGTSAGFFYY67 pKa = 10.66TKK69 pKa = 10.95ANGGDD74 pKa = 3.95DD75 pKa = 4.75DD76 pKa = 6.8DD77 pKa = 6.98GDD79 pKa = 4.34LEE81 pKa = 3.98AARR84 pKa = 11.84AGFYY88 pKa = 10.54FEE90 pKa = 4.49PDD92 pKa = 2.5YY93 pKa = 10.51WYY95 pKa = 10.98YY96 pKa = 11.22YY97 pKa = 9.9IVRR100 pKa = 11.84DD101 pKa = 5.22DD102 pKa = 4.18GDD104 pKa = 4.6LNTGISITPDD114 pKa = 2.97QPADD118 pKa = 3.29NGPPVFHH125 pKa = 6.93GPVPPPPDD133 pKa = 3.78APAPSPPLYY142 pKa = 9.63QCRR145 pKa = 11.84VPGTNFEE152 pKa = 4.25ITFCPSGVWPTDD164 pKa = 3.8DD165 pKa = 6.37DD166 pKa = 5.56NDD168 pKa = 5.34DD169 pKa = 4.05NDD171 pKa = 4.77DD172 pKa = 4.88DD173 pKa = 5.51RR174 pKa = 11.84DD175 pKa = 3.62
MM1 pKa = 7.51VKK3 pKa = 9.59TNPFNFVIAILFASFLHH20 pKa = 6.56VSAQTYY26 pKa = 7.66TVTNRR31 pKa = 11.84CPSAIEE37 pKa = 4.06LFIGQTSEE45 pKa = 4.69GNLATNQSIVKK56 pKa = 8.8TGLGTSAGFFYY67 pKa = 10.66TKK69 pKa = 10.95ANGGDD74 pKa = 3.95DD75 pKa = 4.75DD76 pKa = 6.8DD77 pKa = 6.98GDD79 pKa = 4.34LEE81 pKa = 3.98AARR84 pKa = 11.84AGFYY88 pKa = 10.54FEE90 pKa = 4.49PDD92 pKa = 2.5YY93 pKa = 10.51WYY95 pKa = 10.98YY96 pKa = 11.22YY97 pKa = 9.9IVRR100 pKa = 11.84DD101 pKa = 5.22DD102 pKa = 4.18GDD104 pKa = 4.6LNTGISITPDD114 pKa = 2.97QPADD118 pKa = 3.29NGPPVFHH125 pKa = 6.93GPVPPPPDD133 pKa = 3.78APAPSPPLYY142 pKa = 9.63QCRR145 pKa = 11.84VPGTNFEE152 pKa = 4.25ITFCPSGVWPTDD164 pKa = 3.8DD165 pKa = 6.37DD166 pKa = 5.56NDD168 pKa = 5.34DD169 pKa = 4.05NDD171 pKa = 4.77DD172 pKa = 4.88DD173 pKa = 5.51RR174 pKa = 11.84DD175 pKa = 3.62
Molecular weight: 19.04 kDa
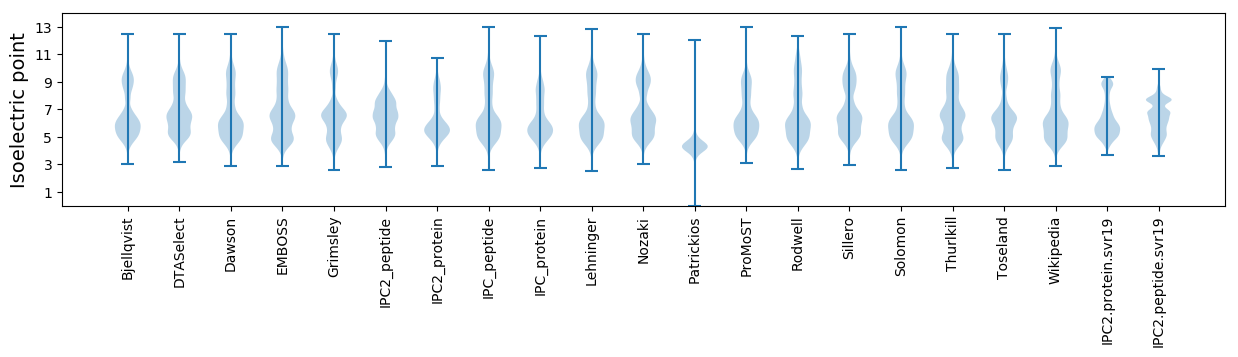
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2DPA6|A0A4Q2DPA6_9AGAR PX domain-containing protein OS=Psathyrella aberdarensis OX=2316362 GN=EST38_g5186 PE=3 SV=1
MM1 pKa = 7.3SGSGGHH7 pKa = 6.02HH8 pKa = 5.7TRR10 pKa = 11.84QGSSTAVKK18 pKa = 10.41VGLHH22 pKa = 4.98GHH24 pKa = 5.79SLNPTPYY31 pKa = 9.01HH32 pKa = 5.78HH33 pKa = 6.97HH34 pKa = 7.14HH35 pKa = 5.93NTGNVTSRR43 pKa = 11.84EE44 pKa = 3.94VVEE47 pKa = 5.22AGNSDD52 pKa = 3.56PRR54 pKa = 11.84SDD56 pKa = 3.5GLAGRR61 pKa = 11.84TPSPTPSEE69 pKa = 3.95LRR71 pKa = 11.84EE72 pKa = 4.2LKK74 pKa = 10.2PAANGWGTSRR84 pKa = 11.84EE85 pKa = 4.07RR86 pKa = 11.84LWFFAAIIILIALIALAWIYY106 pKa = 10.74DD107 pKa = 3.74KK108 pKa = 11.41QIVRR112 pKa = 11.84TLAPAATWLHH122 pKa = 5.84DD123 pKa = 3.76TPGAFAIPMAVLIVISFPPLFGHH146 pKa = 6.77AVVAILCGFVWGLGIGFGIVAGGTFLGEE174 pKa = 3.7IGHH177 pKa = 6.75FYY179 pKa = 10.87ASKK182 pKa = 10.56LVFRR186 pKa = 11.84TRR188 pKa = 11.84GEE190 pKa = 3.85KK191 pKa = 10.55LEE193 pKa = 4.08RR194 pKa = 11.84TNISHH199 pKa = 7.15ACLAQVVRR207 pKa = 11.84DD208 pKa = 3.59GGIRR212 pKa = 11.84ITLIARR218 pKa = 11.84LSAIPAQTTTTVFSTCGVGIIIFAISTLLSMPKK251 pKa = 9.58QALGVYY257 pKa = 10.1LGVILLRR264 pKa = 11.84SRR266 pKa = 11.84FEE268 pKa = 4.23TRR270 pKa = 11.84SSTEE274 pKa = 3.85RR275 pKa = 11.84IVTSVVLAVTMLITAVATWYY295 pKa = 9.9IGRR298 pKa = 11.84QMDD301 pKa = 3.44RR302 pKa = 11.84TKK304 pKa = 10.7PQVIYY309 pKa = 10.47KK310 pKa = 9.87RR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 8.31ARR315 pKa = 11.84QARR318 pKa = 11.84LTQASSPTHH327 pKa = 6.81PYY329 pKa = 11.14LMAASGNGQSTMGSEE344 pKa = 4.32STVMFNPSNDD354 pKa = 3.19NLRR357 pKa = 11.84DD358 pKa = 3.63QNSRR362 pKa = 11.84QAGSSQLALLPPVEE376 pKa = 4.62APQPRR381 pKa = 11.84RR382 pKa = 11.84PFPP385 pKa = 4.1
MM1 pKa = 7.3SGSGGHH7 pKa = 6.02HH8 pKa = 5.7TRR10 pKa = 11.84QGSSTAVKK18 pKa = 10.41VGLHH22 pKa = 4.98GHH24 pKa = 5.79SLNPTPYY31 pKa = 9.01HH32 pKa = 5.78HH33 pKa = 6.97HH34 pKa = 7.14HH35 pKa = 5.93NTGNVTSRR43 pKa = 11.84EE44 pKa = 3.94VVEE47 pKa = 5.22AGNSDD52 pKa = 3.56PRR54 pKa = 11.84SDD56 pKa = 3.5GLAGRR61 pKa = 11.84TPSPTPSEE69 pKa = 3.95LRR71 pKa = 11.84EE72 pKa = 4.2LKK74 pKa = 10.2PAANGWGTSRR84 pKa = 11.84EE85 pKa = 4.07RR86 pKa = 11.84LWFFAAIIILIALIALAWIYY106 pKa = 10.74DD107 pKa = 3.74KK108 pKa = 11.41QIVRR112 pKa = 11.84TLAPAATWLHH122 pKa = 5.84DD123 pKa = 3.76TPGAFAIPMAVLIVISFPPLFGHH146 pKa = 6.77AVVAILCGFVWGLGIGFGIVAGGTFLGEE174 pKa = 3.7IGHH177 pKa = 6.75FYY179 pKa = 10.87ASKK182 pKa = 10.56LVFRR186 pKa = 11.84TRR188 pKa = 11.84GEE190 pKa = 3.85KK191 pKa = 10.55LEE193 pKa = 4.08RR194 pKa = 11.84TNISHH199 pKa = 7.15ACLAQVVRR207 pKa = 11.84DD208 pKa = 3.59GGIRR212 pKa = 11.84ITLIARR218 pKa = 11.84LSAIPAQTTTTVFSTCGVGIIIFAISTLLSMPKK251 pKa = 9.58QALGVYY257 pKa = 10.1LGVILLRR264 pKa = 11.84SRR266 pKa = 11.84FEE268 pKa = 4.23TRR270 pKa = 11.84SSTEE274 pKa = 3.85RR275 pKa = 11.84IVTSVVLAVTMLITAVATWYY295 pKa = 9.9IGRR298 pKa = 11.84QMDD301 pKa = 3.44RR302 pKa = 11.84TKK304 pKa = 10.7PQVIYY309 pKa = 10.47KK310 pKa = 9.87RR311 pKa = 11.84RR312 pKa = 11.84KK313 pKa = 8.31ARR315 pKa = 11.84QARR318 pKa = 11.84LTQASSPTHH327 pKa = 6.81PYY329 pKa = 11.14LMAASGNGQSTMGSEE344 pKa = 4.32STVMFNPSNDD354 pKa = 3.19NLRR357 pKa = 11.84DD358 pKa = 3.63QNSRR362 pKa = 11.84QAGSSQLALLPPVEE376 pKa = 4.62APQPRR381 pKa = 11.84RR382 pKa = 11.84PFPP385 pKa = 4.1
Molecular weight: 41.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6965245 |
10 |
4979 |
474.3 |
52.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.337 ± 0.014 | 1.164 ± 0.006 |
5.792 ± 0.012 | 6.381 ± 0.018 |
3.825 ± 0.011 | 6.422 ± 0.018 |
2.479 ± 0.009 | 4.922 ± 0.013 |
4.981 ± 0.019 | 9.38 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.914 ± 0.007 | 3.632 ± 0.01 |
6.387 ± 0.02 | 3.879 ± 0.013 |
5.972 ± 0.014 | 8.394 ± 0.023 |
5.862 ± 0.014 | 6.186 ± 0.013 |
1.394 ± 0.006 | 2.654 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
