
Dragonfly orbiculatusvirus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.25
Get precalculated fractions of proteins
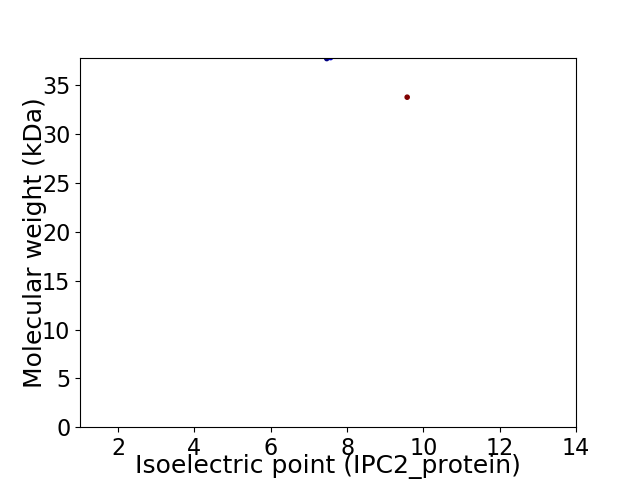
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
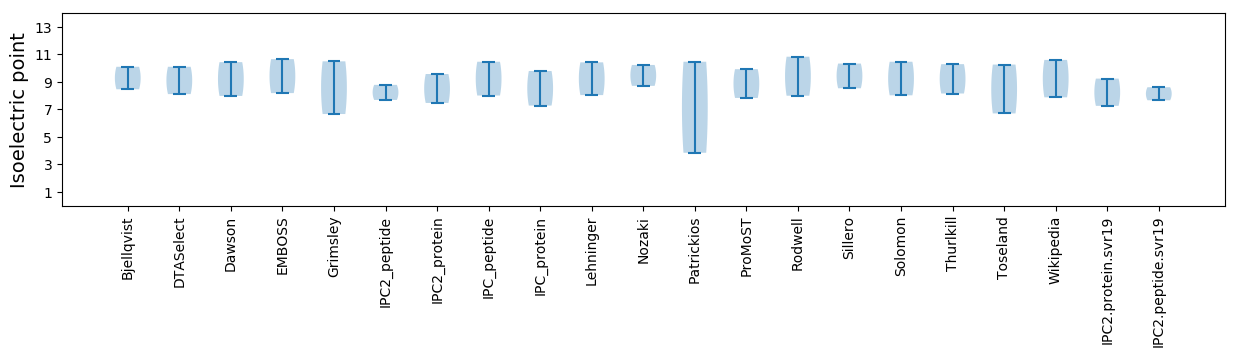
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0A1L1|K0A1L1_9VIRU Replication-associated protein OS=Dragonfly orbiculatusvirus OX=1234874 PE=4 SV=1
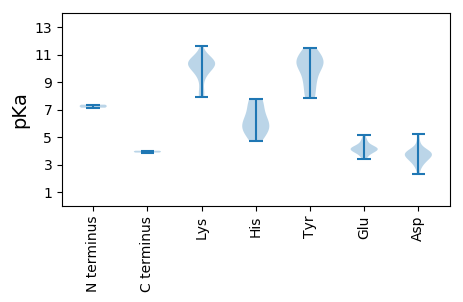
MM1 pKa = 7.13YY2 pKa = 10.38KK3 pKa = 9.99RR4 pKa = 11.84WCFTLNNYY12 pKa = 10.08SVVCNWEE19 pKa = 3.79EE20 pKa = 3.93FFNNGCSTGNKK31 pKa = 7.92LCRR34 pKa = 11.84YY35 pKa = 7.82VVGYY39 pKa = 9.82EE40 pKa = 3.57IGEE43 pKa = 4.23SGTRR47 pKa = 11.84HH48 pKa = 4.69LQGYY52 pKa = 8.64CVLSRR57 pKa = 11.84SQRR60 pKa = 11.84LSYY63 pKa = 8.74VTKK66 pKa = 10.76LFDD69 pKa = 3.64GAHH72 pKa = 5.92WEE74 pKa = 4.26GARR77 pKa = 11.84GSDD80 pKa = 3.33QQNYY84 pKa = 10.15DD85 pKa = 3.38YY86 pKa = 9.35CTKK89 pKa = 10.42SGSFFRR95 pKa = 11.84GGDD98 pKa = 3.3WNVLSRR104 pKa = 11.84SSSGSSDD111 pKa = 3.82RR112 pKa = 11.84IRR114 pKa = 11.84DD115 pKa = 3.81VIKK118 pKa = 9.61TLYY121 pKa = 10.67SMPGSEE127 pKa = 4.62VRR129 pKa = 11.84CSSTYY134 pKa = 10.07VRR136 pKa = 11.84SKK138 pKa = 10.38RR139 pKa = 11.84AIDD142 pKa = 3.68EE143 pKa = 4.27VVSEE147 pKa = 3.94YY148 pKa = 11.07RR149 pKa = 11.84EE150 pKa = 3.89FEE152 pKa = 4.03VRR154 pKa = 11.84KK155 pKa = 9.95LRR157 pKa = 11.84FRR159 pKa = 11.84EE160 pKa = 4.16LSTCLLKK167 pKa = 10.56LWQMSVLLKK176 pKa = 10.79LFNQNDD182 pKa = 3.97RR183 pKa = 11.84QVLWVVDD190 pKa = 3.73VAGGCGKK197 pKa = 8.41TFLSNILFSCYY208 pKa = 10.55SFDD211 pKa = 4.69LFDD214 pKa = 5.2GVTQAKK220 pKa = 9.57DD221 pKa = 2.94ICFLLSEE228 pKa = 4.79TINGIVFDD236 pKa = 3.94VTRR239 pKa = 11.84SDD241 pKa = 3.46SSQFSYY247 pKa = 9.31QTLEE251 pKa = 3.57ACKK254 pKa = 10.28NGFVMTGKK262 pKa = 10.77YY263 pKa = 7.95MGKK266 pKa = 9.17RR267 pKa = 11.84RR268 pKa = 11.84VFKK271 pKa = 10.41SCPVVIFSNCEE282 pKa = 3.4PDD284 pKa = 4.05LVRR287 pKa = 11.84LSSDD291 pKa = 2.3RR292 pKa = 11.84WVVYY296 pKa = 9.16NVPAQARR303 pKa = 11.84SQEE306 pKa = 4.52AIYY309 pKa = 10.32SPQALWPFKK318 pKa = 10.41EE319 pKa = 4.02IEE321 pKa = 4.13AVLLSEE327 pKa = 5.12IEE329 pKa = 4.0
MM1 pKa = 7.13YY2 pKa = 10.38KK3 pKa = 9.99RR4 pKa = 11.84WCFTLNNYY12 pKa = 10.08SVVCNWEE19 pKa = 3.79EE20 pKa = 3.93FFNNGCSTGNKK31 pKa = 7.92LCRR34 pKa = 11.84YY35 pKa = 7.82VVGYY39 pKa = 9.82EE40 pKa = 3.57IGEE43 pKa = 4.23SGTRR47 pKa = 11.84HH48 pKa = 4.69LQGYY52 pKa = 8.64CVLSRR57 pKa = 11.84SQRR60 pKa = 11.84LSYY63 pKa = 8.74VTKK66 pKa = 10.76LFDD69 pKa = 3.64GAHH72 pKa = 5.92WEE74 pKa = 4.26GARR77 pKa = 11.84GSDD80 pKa = 3.33QQNYY84 pKa = 10.15DD85 pKa = 3.38YY86 pKa = 9.35CTKK89 pKa = 10.42SGSFFRR95 pKa = 11.84GGDD98 pKa = 3.3WNVLSRR104 pKa = 11.84SSSGSSDD111 pKa = 3.82RR112 pKa = 11.84IRR114 pKa = 11.84DD115 pKa = 3.81VIKK118 pKa = 9.61TLYY121 pKa = 10.67SMPGSEE127 pKa = 4.62VRR129 pKa = 11.84CSSTYY134 pKa = 10.07VRR136 pKa = 11.84SKK138 pKa = 10.38RR139 pKa = 11.84AIDD142 pKa = 3.68EE143 pKa = 4.27VVSEE147 pKa = 3.94YY148 pKa = 11.07RR149 pKa = 11.84EE150 pKa = 3.89FEE152 pKa = 4.03VRR154 pKa = 11.84KK155 pKa = 9.95LRR157 pKa = 11.84FRR159 pKa = 11.84EE160 pKa = 4.16LSTCLLKK167 pKa = 10.56LWQMSVLLKK176 pKa = 10.79LFNQNDD182 pKa = 3.97RR183 pKa = 11.84QVLWVVDD190 pKa = 3.73VAGGCGKK197 pKa = 8.41TFLSNILFSCYY208 pKa = 10.55SFDD211 pKa = 4.69LFDD214 pKa = 5.2GVTQAKK220 pKa = 9.57DD221 pKa = 2.94ICFLLSEE228 pKa = 4.79TINGIVFDD236 pKa = 3.94VTRR239 pKa = 11.84SDD241 pKa = 3.46SSQFSYY247 pKa = 9.31QTLEE251 pKa = 3.57ACKK254 pKa = 10.28NGFVMTGKK262 pKa = 10.77YY263 pKa = 7.95MGKK266 pKa = 9.17RR267 pKa = 11.84RR268 pKa = 11.84VFKK271 pKa = 10.41SCPVVIFSNCEE282 pKa = 3.4PDD284 pKa = 4.05LVRR287 pKa = 11.84LSSDD291 pKa = 2.3RR292 pKa = 11.84WVVYY296 pKa = 9.16NVPAQARR303 pKa = 11.84SQEE306 pKa = 4.52AIYY309 pKa = 10.32SPQALWPFKK318 pKa = 10.41EE319 pKa = 4.02IEE321 pKa = 4.13AVLLSEE327 pKa = 5.12IEE329 pKa = 4.0
Molecular weight: 37.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0A1L1|K0A1L1_9VIRU Replication-associated protein OS=Dragonfly orbiculatusvirus OX=1234874 PE=4 SV=1
MM1 pKa = 7.35SLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84KK9 pKa = 8.51RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.99IRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.99GRR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84SKK25 pKa = 9.29RR26 pKa = 11.84SCFPRR31 pKa = 11.84LNNFYY36 pKa = 11.17SKK38 pKa = 9.9MCIKK42 pKa = 10.38EE43 pKa = 4.07VTQFTATSNATHH55 pKa = 5.53VHH57 pKa = 6.32SISLANVNSYY67 pKa = 10.37VLQFLQNFKK76 pKa = 10.27YY77 pKa = 10.14YY78 pKa = 10.76RR79 pKa = 11.84MYY81 pKa = 11.44AMSVLLMPRR90 pKa = 11.84MNAIGATQSYY100 pKa = 7.89YY101 pKa = 10.61QPQNDD106 pKa = 3.06AHH108 pKa = 7.15LINTAPNLWTLFEE121 pKa = 4.84PEE123 pKa = 4.05TGEE126 pKa = 4.2VNDD129 pKa = 3.72VVALHH134 pKa = 5.89LHH136 pKa = 5.57PRR138 pKa = 11.84SRR140 pKa = 11.84LHH142 pKa = 6.62SFNKK146 pKa = 8.01TVRR149 pKa = 11.84RR150 pKa = 11.84FIKK153 pKa = 9.69LQPSLRR159 pKa = 11.84VNTADD164 pKa = 3.99LSNVDD169 pKa = 2.75IHH171 pKa = 7.69GGVFRR176 pKa = 11.84KK177 pKa = 7.89TWVAMADD184 pKa = 4.3FKK186 pKa = 11.63ASFGKK191 pKa = 10.11FVMAFSEE198 pKa = 4.44DD199 pKa = 3.45TKK201 pKa = 11.13SDD203 pKa = 3.71QNIHH207 pKa = 5.26TEE209 pKa = 3.8YY210 pKa = 11.22DD211 pKa = 3.69VVFKK215 pKa = 10.39YY216 pKa = 11.19YY217 pKa = 10.43IMLKK221 pKa = 10.54GSAPNLWSGEE231 pKa = 4.15VVTQQLPAPEE241 pKa = 4.38TVPLASSKK249 pKa = 10.93GGSVKK254 pKa = 10.38KK255 pKa = 10.54KK256 pKa = 10.44SGDD259 pKa = 3.18IPVIKK264 pKa = 10.16VRR266 pKa = 11.84QVDD269 pKa = 3.79TGPVPDD275 pKa = 4.0SHH277 pKa = 7.74GLGQKK282 pKa = 9.92SSAIEE287 pKa = 3.91KK288 pKa = 10.21VLEE291 pKa = 4.15EE292 pKa = 4.18EE293 pKa = 4.26MGLL296 pKa = 3.87
MM1 pKa = 7.35SLRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GRR8 pKa = 11.84KK9 pKa = 8.51RR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 7.99IRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84YY18 pKa = 8.99GRR20 pKa = 11.84SRR22 pKa = 11.84RR23 pKa = 11.84SKK25 pKa = 9.29RR26 pKa = 11.84SCFPRR31 pKa = 11.84LNNFYY36 pKa = 11.17SKK38 pKa = 9.9MCIKK42 pKa = 10.38EE43 pKa = 4.07VTQFTATSNATHH55 pKa = 5.53VHH57 pKa = 6.32SISLANVNSYY67 pKa = 10.37VLQFLQNFKK76 pKa = 10.27YY77 pKa = 10.14YY78 pKa = 10.76RR79 pKa = 11.84MYY81 pKa = 11.44AMSVLLMPRR90 pKa = 11.84MNAIGATQSYY100 pKa = 7.89YY101 pKa = 10.61QPQNDD106 pKa = 3.06AHH108 pKa = 7.15LINTAPNLWTLFEE121 pKa = 4.84PEE123 pKa = 4.05TGEE126 pKa = 4.2VNDD129 pKa = 3.72VVALHH134 pKa = 5.89LHH136 pKa = 5.57PRR138 pKa = 11.84SRR140 pKa = 11.84LHH142 pKa = 6.62SFNKK146 pKa = 8.01TVRR149 pKa = 11.84RR150 pKa = 11.84FIKK153 pKa = 9.69LQPSLRR159 pKa = 11.84VNTADD164 pKa = 3.99LSNVDD169 pKa = 2.75IHH171 pKa = 7.69GGVFRR176 pKa = 11.84KK177 pKa = 7.89TWVAMADD184 pKa = 4.3FKK186 pKa = 11.63ASFGKK191 pKa = 10.11FVMAFSEE198 pKa = 4.44DD199 pKa = 3.45TKK201 pKa = 11.13SDD203 pKa = 3.71QNIHH207 pKa = 5.26TEE209 pKa = 3.8YY210 pKa = 11.22DD211 pKa = 3.69VVFKK215 pKa = 10.39YY216 pKa = 11.19YY217 pKa = 10.43IMLKK221 pKa = 10.54GSAPNLWSGEE231 pKa = 4.15VVTQQLPAPEE241 pKa = 4.38TVPLASSKK249 pKa = 10.93GGSVKK254 pKa = 10.38KK255 pKa = 10.54KK256 pKa = 10.44SGDD259 pKa = 3.18IPVIKK264 pKa = 10.16VRR266 pKa = 11.84QVDD269 pKa = 3.79TGPVPDD275 pKa = 4.0SHH277 pKa = 7.74GLGQKK282 pKa = 9.92SSAIEE287 pKa = 3.91KK288 pKa = 10.21VLEE291 pKa = 4.15EE292 pKa = 4.18EE293 pKa = 4.26MGLL296 pKa = 3.87
Molecular weight: 33.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
625 |
296 |
329 |
312.5 |
35.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.64 ± 1.016 | 2.56 ± 1.328 |
4.32 ± 0.426 | 4.96 ± 0.639 |
5.44 ± 0.501 | 6.08 ± 0.475 |
1.76 ± 0.903 | 3.84 ± 0.151 |
5.76 ± 0.703 | 8.16 ± 0.275 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.4 ± 0.69 | 4.96 ± 0.314 |
3.2 ± 1.078 | 4.0 ± 0.038 |
7.68 ± 0.302 | 10.4 ± 0.901 |
4.8 ± 0.427 | 8.8 ± 0.25 |
1.76 ± 0.526 | 4.48 ± 0.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
