
Chimpanzee polyomavirus Bob
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus; Pan troglodytes polyomavirus 1
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
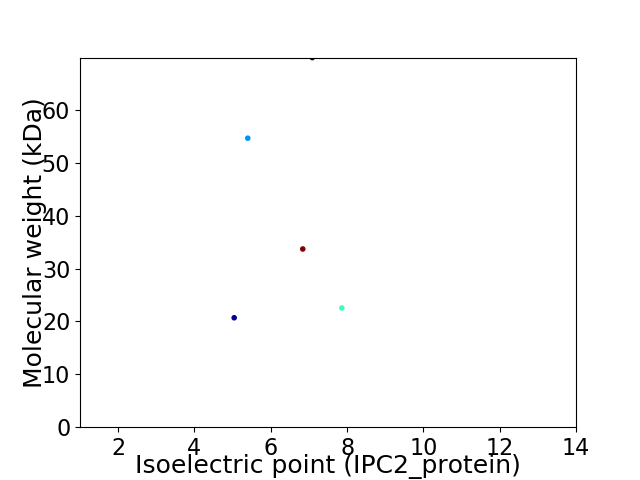
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E5AX30|E5AX30_9POLY VP1 protein OS=Chimpanzee polyomavirus Bob OX=2035845 GN=VP1 PE=3 SV=1
MM1 pKa = 7.01TVEE4 pKa = 4.18GLSGIEE10 pKa = 4.47ALAQLGWTAEE20 pKa = 4.19QFSNMAFISTTFSNAIGYY38 pKa = 8.57GVLFQTVSGISSLVSAGIRR57 pKa = 11.84LGTSVSSVNRR67 pKa = 11.84HH68 pKa = 3.58QTEE71 pKa = 4.1QEE73 pKa = 3.93LEE75 pKa = 4.16TLFGKK80 pKa = 9.98IAHH83 pKa = 6.47FLHH86 pKa = 6.58VNLAFHH92 pKa = 7.44LDD94 pKa = 3.46PFDD97 pKa = 3.35WCGSIGTTMPPEE109 pKa = 4.47FSNLTLDD116 pKa = 3.85QLSKK120 pKa = 10.81LALIIEE126 pKa = 4.27NGRR129 pKa = 11.84WVIQRR134 pKa = 11.84SPTHH138 pKa = 7.03DD139 pKa = 3.82PLFEE143 pKa = 6.25SGDD146 pKa = 3.89IIDD149 pKa = 4.1MFGPPGGARR158 pKa = 11.84QRR160 pKa = 11.84VTPDD164 pKa = 2.19WMLPLILRR172 pKa = 11.84LNGASQEE179 pKa = 4.07KK180 pKa = 10.73SSLCVNSNQSS190 pKa = 2.84
MM1 pKa = 7.01TVEE4 pKa = 4.18GLSGIEE10 pKa = 4.47ALAQLGWTAEE20 pKa = 4.19QFSNMAFISTTFSNAIGYY38 pKa = 8.57GVLFQTVSGISSLVSAGIRR57 pKa = 11.84LGTSVSSVNRR67 pKa = 11.84HH68 pKa = 3.58QTEE71 pKa = 4.1QEE73 pKa = 3.93LEE75 pKa = 4.16TLFGKK80 pKa = 9.98IAHH83 pKa = 6.47FLHH86 pKa = 6.58VNLAFHH92 pKa = 7.44LDD94 pKa = 3.46PFDD97 pKa = 3.35WCGSIGTTMPPEE109 pKa = 4.47FSNLTLDD116 pKa = 3.85QLSKK120 pKa = 10.81LALIIEE126 pKa = 4.27NGRR129 pKa = 11.84WVIQRR134 pKa = 11.84SPTHH138 pKa = 7.03DD139 pKa = 3.82PLFEE143 pKa = 6.25SGDD146 pKa = 3.89IIDD149 pKa = 4.1MFGPPGGARR158 pKa = 11.84QRR160 pKa = 11.84VTPDD164 pKa = 2.19WMLPLILRR172 pKa = 11.84LNGASQEE179 pKa = 4.07KK180 pKa = 10.73SSLCVNSNQSS190 pKa = 2.84
Molecular weight: 20.7 kDa
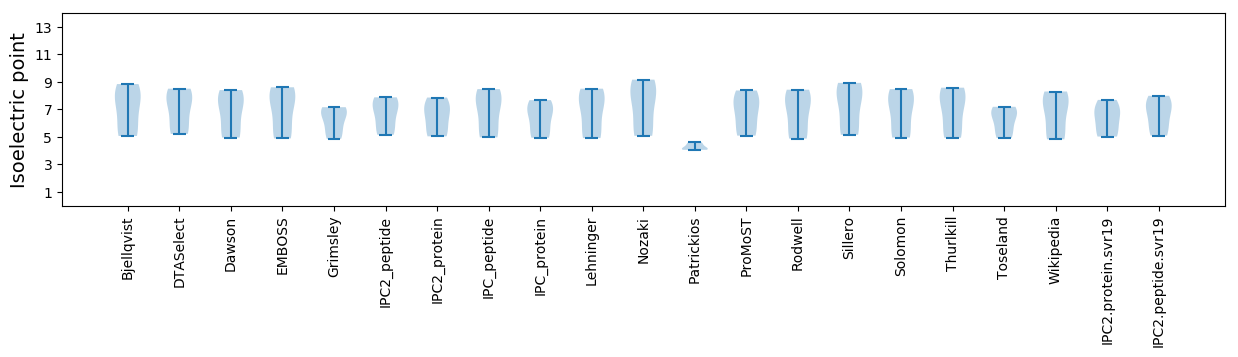
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E5AX32|E5AX32_9POLY Small T antigen OS=Chimpanzee polyomavirus Bob OX=2035845 GN=small t PE=4 SV=1
MM1 pKa = 7.97DD2 pKa = 4.63KK3 pKa = 10.86VLEE6 pKa = 4.33KK7 pKa = 10.41SDD9 pKa = 4.2RR10 pKa = 11.84EE11 pKa = 4.03MLIEE15 pKa = 3.95LLGIPSYY22 pKa = 11.62AFGNFPIMKK31 pKa = 8.51TAYY34 pKa = 9.07KK35 pKa = 9.52RR36 pKa = 11.84ASKK39 pKa = 9.97IYY41 pKa = 10.0HH42 pKa = 6.58PDD44 pKa = 3.14KK45 pKa = 11.07GGSSEE50 pKa = 5.93KK51 pKa = 10.35MMLLNSLWQKK61 pKa = 9.64FQEE64 pKa = 4.29GLVDD68 pKa = 3.31IRR70 pKa = 11.84GSEE73 pKa = 4.16VCQVSFSDD81 pKa = 4.43CYY83 pKa = 10.86DD84 pKa = 3.17VKK86 pKa = 11.01LLRR89 pKa = 11.84NCGTVKK95 pKa = 10.27HH96 pKa = 5.46FHH98 pKa = 6.72EE99 pKa = 4.89IFLRR103 pKa = 11.84SPQCLQKK110 pKa = 11.06GSAVCNCITSTLFNQHH126 pKa = 5.47RR127 pKa = 11.84QIKK130 pKa = 8.61LLCNKK135 pKa = 9.81RR136 pKa = 11.84CLTWGEE142 pKa = 4.54CFCFSCFLIWFGMDD156 pKa = 3.83FKK158 pKa = 11.02WEE160 pKa = 4.09SFDD163 pKa = 2.97MWKK166 pKa = 10.58YY167 pKa = 10.84VIAEE171 pKa = 4.31MPTGLLQLPPSKK183 pKa = 10.97YY184 pKa = 9.86KK185 pKa = 10.6FSSFPLVLKK194 pKa = 10.77
MM1 pKa = 7.97DD2 pKa = 4.63KK3 pKa = 10.86VLEE6 pKa = 4.33KK7 pKa = 10.41SDD9 pKa = 4.2RR10 pKa = 11.84EE11 pKa = 4.03MLIEE15 pKa = 3.95LLGIPSYY22 pKa = 11.62AFGNFPIMKK31 pKa = 8.51TAYY34 pKa = 9.07KK35 pKa = 9.52RR36 pKa = 11.84ASKK39 pKa = 9.97IYY41 pKa = 10.0HH42 pKa = 6.58PDD44 pKa = 3.14KK45 pKa = 11.07GGSSEE50 pKa = 5.93KK51 pKa = 10.35MMLLNSLWQKK61 pKa = 9.64FQEE64 pKa = 4.29GLVDD68 pKa = 3.31IRR70 pKa = 11.84GSEE73 pKa = 4.16VCQVSFSDD81 pKa = 4.43CYY83 pKa = 10.86DD84 pKa = 3.17VKK86 pKa = 11.01LLRR89 pKa = 11.84NCGTVKK95 pKa = 10.27HH96 pKa = 5.46FHH98 pKa = 6.72EE99 pKa = 4.89IFLRR103 pKa = 11.84SPQCLQKK110 pKa = 11.06GSAVCNCITSTLFNQHH126 pKa = 5.47RR127 pKa = 11.84QIKK130 pKa = 8.61LLCNKK135 pKa = 9.81RR136 pKa = 11.84CLTWGEE142 pKa = 4.54CFCFSCFLIWFGMDD156 pKa = 3.83FKK158 pKa = 11.02WEE160 pKa = 4.09SFDD163 pKa = 2.97MWKK166 pKa = 10.58YY167 pKa = 10.84VIAEE171 pKa = 4.31MPTGLLQLPPSKK183 pKa = 10.97YY184 pKa = 9.86KK185 pKa = 10.6FSSFPLVLKK194 pKa = 10.77
Molecular weight: 22.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1805 |
190 |
615 |
361.0 |
40.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.427 ± 0.515 | 2.382 ± 0.439 |
4.598 ± 0.502 | 6.371 ± 0.502 |
5.097 ± 0.809 | 6.593 ± 0.852 |
2.05 ± 0.117 | 5.762 ± 0.242 |
6.482 ± 1.273 | 10.693 ± 0.892 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.992 ± 0.154 | 4.155 ± 0.437 |
5.817 ± 1.374 | 4.321 ± 0.258 |
4.1 ± 0.369 | 7.756 ± 1.141 |
5.485 ± 0.807 | 5.263 ± 0.313 |
1.33 ± 0.274 | 2.327 ± 0.548 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
