
Telmatospirillum siberiense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Telmatospirillum
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
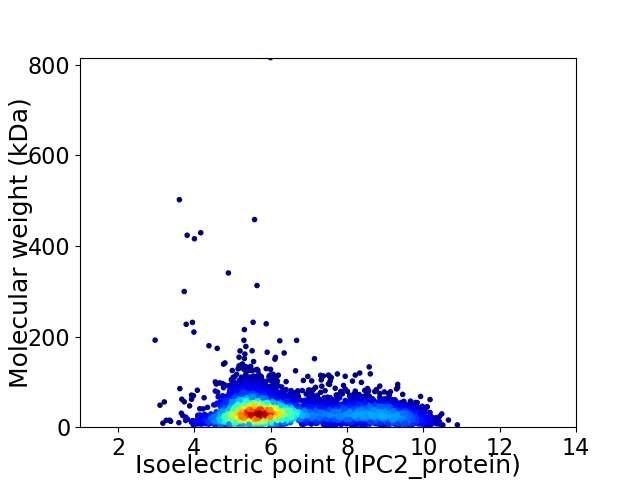
Virtual 2D-PAGE plot for 5353 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N3PM31|A0A2N3PM31_9PROT Uncharacterized protein OS=Telmatospirillum siberiense OX=382514 GN=CWS72_26640 PE=4 SV=1
MM1 pKa = 6.58QQASIDD7 pKa = 4.22AIIAAAKK14 pKa = 10.59ASDD17 pKa = 4.55LDD19 pKa = 4.46LNDD22 pKa = 3.49TALLLSIIRR31 pKa = 11.84TEE33 pKa = 4.39SGFNPNAAAGSSSASGLGQFINGTAAQYY61 pKa = 11.76GLTASNRR68 pKa = 11.84WSIAAQAQAAVDD80 pKa = 4.26LFIDD84 pKa = 4.24CQEE87 pKa = 4.07KK88 pKa = 10.95AGDD91 pKa = 3.68NGQDD95 pKa = 3.07SSHH98 pKa = 7.14IYY100 pKa = 10.31AYY102 pKa = 9.24WHH104 pKa = 7.41DD105 pKa = 3.85GLGTDD110 pKa = 5.35GYY112 pKa = 11.49GAGLKK117 pKa = 9.21TSNASVMPNVSAFTQALVAGNYY139 pKa = 9.56GLASSSTPISPDD151 pKa = 3.1TLTTNVDD158 pKa = 3.03GTTTLTRR165 pKa = 11.84VSNGSTYY172 pKa = 10.94SEE174 pKa = 4.26TFSAGGAANLSVTTNGQVTSSDD196 pKa = 3.52YY197 pKa = 11.16DD198 pKa = 3.47SSGRR202 pKa = 11.84LVSSATSGTSDD213 pKa = 3.32SFTAGGNTIAISGAQSLLSQGAGGYY238 pKa = 10.12SLTDD242 pKa = 3.29ASGNIYY248 pKa = 8.8TLSQLNDD255 pKa = 3.04GSGQYY260 pKa = 10.9RR261 pKa = 11.84LTSSSGLINVTLDD274 pKa = 3.23PSIVTDD280 pKa = 3.43IQLGANLKK288 pKa = 9.93IDD290 pKa = 3.52TALGDD295 pKa = 3.71FVTINTQFGSGAFGFVGSSQTAFNSPSLINIDD327 pKa = 3.21QNYY330 pKa = 8.26FQLGTPNGGLGATYY344 pKa = 9.71DD345 pKa = 3.59IQPNGSLGATIAGFLGLSGSTDD367 pKa = 3.58FASLLNEE374 pKa = 4.39TSALLPSLTAAVPTDD389 pKa = 3.63APVSVPTFSKK399 pKa = 9.12TALGVTVTDD408 pKa = 4.79LATAAQTAAGSGGDD422 pKa = 3.44GTAYY426 pKa = 10.79SITNAIWDD434 pKa = 4.17TASSLITEE442 pKa = 5.21AGQALTDD449 pKa = 3.78VADD452 pKa = 4.03VSASEE457 pKa = 4.16IANALWRR464 pKa = 11.84PYY466 pKa = 8.97YY467 pKa = 10.07QSLMSYY473 pKa = 10.6FNDD476 pKa = 3.1VPAATGVLNTDD487 pKa = 3.4AGSVLLPGGSGVTVGDD503 pKa = 4.04LVAAAVDD510 pKa = 4.36SNPTPSFTDD519 pKa = 3.63PLLLDD524 pKa = 3.85LTGGGIGVSSWIRR537 pKa = 11.84SPVYY541 pKa = 10.29FDD543 pKa = 4.01TNVKK547 pKa = 10.42ADD549 pKa = 3.65ANGNPTTTPDD559 pKa = 3.43GMEE562 pKa = 5.11HH563 pKa = 5.03EE564 pKa = 4.65TAWMKK569 pKa = 10.92AGTGMLVFEE578 pKa = 4.85SGGVVAPITNITQTVSEE595 pKa = 4.64FLNAGATPGKK605 pKa = 10.37YY606 pKa = 10.04ADD608 pKa = 4.3GLAALL613 pKa = 4.73
MM1 pKa = 6.58QQASIDD7 pKa = 4.22AIIAAAKK14 pKa = 10.59ASDD17 pKa = 4.55LDD19 pKa = 4.46LNDD22 pKa = 3.49TALLLSIIRR31 pKa = 11.84TEE33 pKa = 4.39SGFNPNAAAGSSSASGLGQFINGTAAQYY61 pKa = 11.76GLTASNRR68 pKa = 11.84WSIAAQAQAAVDD80 pKa = 4.26LFIDD84 pKa = 4.24CQEE87 pKa = 4.07KK88 pKa = 10.95AGDD91 pKa = 3.68NGQDD95 pKa = 3.07SSHH98 pKa = 7.14IYY100 pKa = 10.31AYY102 pKa = 9.24WHH104 pKa = 7.41DD105 pKa = 3.85GLGTDD110 pKa = 5.35GYY112 pKa = 11.49GAGLKK117 pKa = 9.21TSNASVMPNVSAFTQALVAGNYY139 pKa = 9.56GLASSSTPISPDD151 pKa = 3.1TLTTNVDD158 pKa = 3.03GTTTLTRR165 pKa = 11.84VSNGSTYY172 pKa = 10.94SEE174 pKa = 4.26TFSAGGAANLSVTTNGQVTSSDD196 pKa = 3.52YY197 pKa = 11.16DD198 pKa = 3.47SSGRR202 pKa = 11.84LVSSATSGTSDD213 pKa = 3.32SFTAGGNTIAISGAQSLLSQGAGGYY238 pKa = 10.12SLTDD242 pKa = 3.29ASGNIYY248 pKa = 8.8TLSQLNDD255 pKa = 3.04GSGQYY260 pKa = 10.9RR261 pKa = 11.84LTSSSGLINVTLDD274 pKa = 3.23PSIVTDD280 pKa = 3.43IQLGANLKK288 pKa = 9.93IDD290 pKa = 3.52TALGDD295 pKa = 3.71FVTINTQFGSGAFGFVGSSQTAFNSPSLINIDD327 pKa = 3.21QNYY330 pKa = 8.26FQLGTPNGGLGATYY344 pKa = 9.71DD345 pKa = 3.59IQPNGSLGATIAGFLGLSGSTDD367 pKa = 3.58FASLLNEE374 pKa = 4.39TSALLPSLTAAVPTDD389 pKa = 3.63APVSVPTFSKK399 pKa = 9.12TALGVTVTDD408 pKa = 4.79LATAAQTAAGSGGDD422 pKa = 3.44GTAYY426 pKa = 10.79SITNAIWDD434 pKa = 4.17TASSLITEE442 pKa = 5.21AGQALTDD449 pKa = 3.78VADD452 pKa = 4.03VSASEE457 pKa = 4.16IANALWRR464 pKa = 11.84PYY466 pKa = 8.97YY467 pKa = 10.07QSLMSYY473 pKa = 10.6FNDD476 pKa = 3.1VPAATGVLNTDD487 pKa = 3.4AGSVLLPGGSGVTVGDD503 pKa = 4.04LVAAAVDD510 pKa = 4.36SNPTPSFTDD519 pKa = 3.63PLLLDD524 pKa = 3.85LTGGGIGVSSWIRR537 pKa = 11.84SPVYY541 pKa = 10.29FDD543 pKa = 4.01TNVKK547 pKa = 10.42ADD549 pKa = 3.65ANGNPTTTPDD559 pKa = 3.43GMEE562 pKa = 5.11HH563 pKa = 5.03EE564 pKa = 4.65TAWMKK569 pKa = 10.92AGTGMLVFEE578 pKa = 4.85SGGVVAPITNITQTVSEE595 pKa = 4.64FLNAGATPGKK605 pKa = 10.37YY606 pKa = 10.04ADD608 pKa = 4.3GLAALL613 pKa = 4.73
Molecular weight: 61.68 kDa
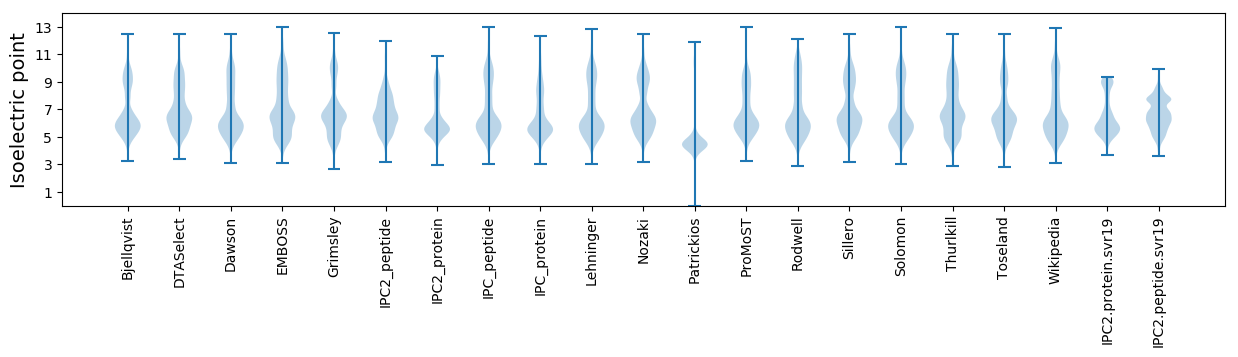
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N3PRL2|A0A2N3PRL2_9PROT Uncharacterized protein OS=Telmatospirillum siberiense OX=382514 GN=CWS72_18540 PE=4 SV=1
MM1 pKa = 7.3TSAALPARR9 pKa = 11.84AGSRR13 pKa = 11.84TLQSIARR20 pKa = 11.84IGSAATLLILFAGIGSTVGWPGLRR44 pKa = 11.84FDD46 pKa = 5.49FLSQHH51 pKa = 6.4SGFQISEE58 pKa = 4.18SLFQIGADD66 pKa = 3.52APIWLMLIAGLINTIRR82 pKa = 11.84VSAMAMVLTIPLGIALALLRR102 pKa = 11.84GSAFRR107 pKa = 11.84PGRR110 pKa = 11.84WLATGVIEE118 pKa = 5.4PIRR121 pKa = 11.84NTPVLLQLFLWYY133 pKa = 10.53GIVTQLLPGPRR144 pKa = 11.84QAVEE148 pKa = 4.04IPPGILLCNRR158 pKa = 11.84GLYY161 pKa = 9.74FPWFDD166 pKa = 3.29GSGVTWPKK174 pKa = 10.35LAGFNIEE181 pKa = 4.4GGLSISPEE189 pKa = 3.77FTVLVFGLALFHH201 pKa = 6.45GCYY204 pKa = 9.56LAEE207 pKa = 4.13IFRR210 pKa = 11.84AGFLAVPRR218 pKa = 11.84GQRR221 pKa = 11.84DD222 pKa = 3.21AAKK225 pKa = 9.56ALSMPAFSTLRR236 pKa = 11.84KK237 pKa = 9.37ILLPQALAFALPSTAAQILALVKK260 pKa = 10.73NSSLAIAIGFADD272 pKa = 5.32FVAVLNTSVNQNGRR286 pKa = 11.84TLEE289 pKa = 3.93GLIVIILVFLGLNTLLGAAVEE310 pKa = 4.22KK311 pKa = 9.67GTRR314 pKa = 11.84RR315 pKa = 11.84FGKK318 pKa = 8.75ATRR321 pKa = 11.84GQLAVEE327 pKa = 4.19RR328 pKa = 11.84QSMEE332 pKa = 3.72DD333 pKa = 3.18RR334 pKa = 11.84KK335 pKa = 10.01PAGLRR340 pKa = 11.84VCSDD344 pKa = 2.6IAMIATATAIFLPAIVRR361 pKa = 11.84IFRR364 pKa = 11.84WGVLDD369 pKa = 4.05AAWAGSPADD378 pKa = 3.34CAMANGACWAVVTEE392 pKa = 4.45KK393 pKa = 11.17APLLLFGPFPGAEE406 pKa = 3.72TWRR409 pKa = 11.84PLIATLLIILFIIALGIDD427 pKa = 3.87RR428 pKa = 11.84LRR430 pKa = 11.84HH431 pKa = 5.65AGVSLAALAATMIGWIWLLGGGAGLASVPTNAWGGLSLTSGLAVAVVMGACLLALPLALARR492 pKa = 11.84RR493 pKa = 11.84SPHH496 pKa = 5.18KK497 pKa = 9.18TIHH500 pKa = 5.67IPAVIFIEE508 pKa = 4.16IFRR511 pKa = 11.84GVPLVALLLSADD523 pKa = 3.35ILIPGLLPSGWQFDD537 pKa = 4.34KK538 pKa = 11.04LWRR541 pKa = 11.84AYY543 pKa = 10.28AAITLLASVNLAEE556 pKa = 4.44VMRR559 pKa = 11.84GALAAVAPGQTEE571 pKa = 3.8AARR574 pKa = 11.84ALGLSARR581 pKa = 11.84RR582 pKa = 11.84SFLLVILPQAIRR594 pKa = 11.84IAMPALVNVFVGAIKK609 pKa = 9.5DD610 pKa = 3.61TSLVLIVGILDD621 pKa = 3.1ITGAAKK627 pKa = 10.3AAVADD632 pKa = 3.97PAWRR636 pKa = 11.84NFAPEE641 pKa = 3.89IYY643 pKa = 10.28LFLAAVYY650 pKa = 8.78FALCFPIGRR659 pKa = 11.84LARR662 pKa = 11.84RR663 pKa = 11.84MTDD666 pKa = 2.72QTTADD671 pKa = 3.48SPSRR675 pKa = 11.84KK676 pKa = 9.18RR677 pKa = 11.84SISAASLL684 pKa = 3.4
MM1 pKa = 7.3TSAALPARR9 pKa = 11.84AGSRR13 pKa = 11.84TLQSIARR20 pKa = 11.84IGSAATLLILFAGIGSTVGWPGLRR44 pKa = 11.84FDD46 pKa = 5.49FLSQHH51 pKa = 6.4SGFQISEE58 pKa = 4.18SLFQIGADD66 pKa = 3.52APIWLMLIAGLINTIRR82 pKa = 11.84VSAMAMVLTIPLGIALALLRR102 pKa = 11.84GSAFRR107 pKa = 11.84PGRR110 pKa = 11.84WLATGVIEE118 pKa = 5.4PIRR121 pKa = 11.84NTPVLLQLFLWYY133 pKa = 10.53GIVTQLLPGPRR144 pKa = 11.84QAVEE148 pKa = 4.04IPPGILLCNRR158 pKa = 11.84GLYY161 pKa = 9.74FPWFDD166 pKa = 3.29GSGVTWPKK174 pKa = 10.35LAGFNIEE181 pKa = 4.4GGLSISPEE189 pKa = 3.77FTVLVFGLALFHH201 pKa = 6.45GCYY204 pKa = 9.56LAEE207 pKa = 4.13IFRR210 pKa = 11.84AGFLAVPRR218 pKa = 11.84GQRR221 pKa = 11.84DD222 pKa = 3.21AAKK225 pKa = 9.56ALSMPAFSTLRR236 pKa = 11.84KK237 pKa = 9.37ILLPQALAFALPSTAAQILALVKK260 pKa = 10.73NSSLAIAIGFADD272 pKa = 5.32FVAVLNTSVNQNGRR286 pKa = 11.84TLEE289 pKa = 3.93GLIVIILVFLGLNTLLGAAVEE310 pKa = 4.22KK311 pKa = 9.67GTRR314 pKa = 11.84RR315 pKa = 11.84FGKK318 pKa = 8.75ATRR321 pKa = 11.84GQLAVEE327 pKa = 4.19RR328 pKa = 11.84QSMEE332 pKa = 3.72DD333 pKa = 3.18RR334 pKa = 11.84KK335 pKa = 10.01PAGLRR340 pKa = 11.84VCSDD344 pKa = 2.6IAMIATATAIFLPAIVRR361 pKa = 11.84IFRR364 pKa = 11.84WGVLDD369 pKa = 4.05AAWAGSPADD378 pKa = 3.34CAMANGACWAVVTEE392 pKa = 4.45KK393 pKa = 11.17APLLLFGPFPGAEE406 pKa = 3.72TWRR409 pKa = 11.84PLIATLLIILFIIALGIDD427 pKa = 3.87RR428 pKa = 11.84LRR430 pKa = 11.84HH431 pKa = 5.65AGVSLAALAATMIGWIWLLGGGAGLASVPTNAWGGLSLTSGLAVAVVMGACLLALPLALARR492 pKa = 11.84RR493 pKa = 11.84SPHH496 pKa = 5.18KK497 pKa = 9.18TIHH500 pKa = 5.67IPAVIFIEE508 pKa = 4.16IFRR511 pKa = 11.84GVPLVALLLSADD523 pKa = 3.35ILIPGLLPSGWQFDD537 pKa = 4.34KK538 pKa = 11.04LWRR541 pKa = 11.84AYY543 pKa = 10.28AAITLLASVNLAEE556 pKa = 4.44VMRR559 pKa = 11.84GALAAVAPGQTEE571 pKa = 3.8AARR574 pKa = 11.84ALGLSARR581 pKa = 11.84RR582 pKa = 11.84SFLLVILPQAIRR594 pKa = 11.84IAMPALVNVFVGAIKK609 pKa = 9.5DD610 pKa = 3.61TSLVLIVGILDD621 pKa = 3.1ITGAAKK627 pKa = 10.3AAVADD632 pKa = 3.97PAWRR636 pKa = 11.84NFAPEE641 pKa = 3.89IYY643 pKa = 10.28LFLAAVYY650 pKa = 8.78FALCFPIGRR659 pKa = 11.84LARR662 pKa = 11.84RR663 pKa = 11.84MTDD666 pKa = 2.72QTTADD671 pKa = 3.48SPSRR675 pKa = 11.84KK676 pKa = 9.18RR677 pKa = 11.84SISAASLL684 pKa = 3.4
Molecular weight: 72.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1820509 |
26 |
7446 |
340.1 |
36.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.178 ± 0.051 | 0.951 ± 0.013 |
5.6 ± 0.026 | 5.362 ± 0.033 |
3.626 ± 0.025 | 8.73 ± 0.042 |
2.085 ± 0.017 | 5.189 ± 0.025 |
3.102 ± 0.025 | 10.695 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.336 ± 0.017 | 2.586 ± 0.022 |
5.119 ± 0.031 | 3.178 ± 0.022 |
7.019 ± 0.049 | 5.841 ± 0.041 |
5.425 ± 0.058 | 7.507 ± 0.029 |
1.309 ± 0.016 | 2.16 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
