
Cytophagaceae bacterium SJW1-29
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cytophagaceae; unclassified Cytophagaceae
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
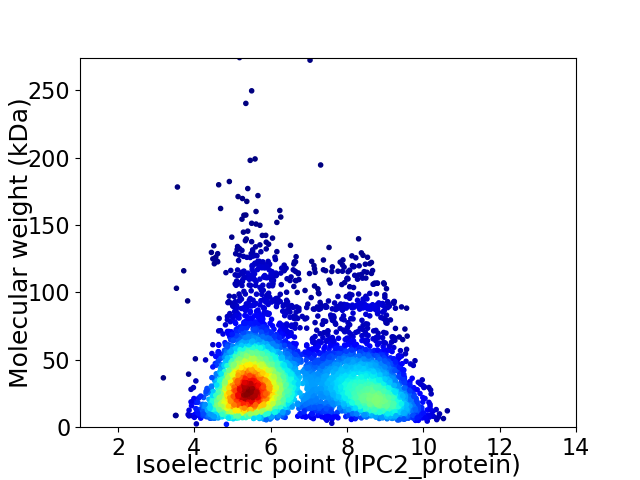
Virtual 2D-PAGE plot for 5629 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7C9B9M3|A0A7C9B9M3_9BACT DinB family protein OS=Cytophagaceae bacterium SJW1-29 OX=2654236 GN=GBK04_08815 PE=4 SV=1
MM1 pKa = 7.41VDD3 pKa = 3.38GVTVAVVNSDD13 pKa = 3.14STLTIKK19 pKa = 11.01GPGSYY24 pKa = 9.09TFLSTSGVLYY34 pKa = 9.42PASGFCAIEE43 pKa = 4.07VVPGAFFDD51 pKa = 4.22LALSKK56 pKa = 10.9SLAPGQASTVTAGSLVKK73 pKa = 10.28FRR75 pKa = 11.84LTVSNEE81 pKa = 3.59GNMEE85 pKa = 4.24ALQIALSDD93 pKa = 3.86SLPEE97 pKa = 4.23GMTLADD103 pKa = 4.02TSWTASGGFATLNVPLAGPLYY124 pKa = 9.86PGATAFIDD132 pKa = 3.03ITLQVDD138 pKa = 3.42SSFTSGTLTNFAQIKK153 pKa = 9.52DD154 pKa = 3.77ASCATDD160 pKa = 3.78EE161 pKa = 4.29PVIDD165 pKa = 4.86IDD167 pKa = 3.92STPDD171 pKa = 2.93NGFNNGEE178 pKa = 4.46DD179 pKa = 4.13DD180 pKa = 5.27DD181 pKa = 5.54DD182 pKa = 5.46SEE184 pKa = 5.41SISISQCPTITLTTSSDD201 pKa = 3.2KK202 pKa = 11.03VICEE206 pKa = 4.23GEE208 pKa = 4.45SVVLNVTASEE218 pKa = 4.21SGATIDD224 pKa = 3.79WYY226 pKa = 9.52TVPTGGTPFVTTASEE241 pKa = 4.12ADD243 pKa = 3.47QTLSPTQTTTYY254 pKa = 9.61YY255 pKa = 10.92VEE257 pKa = 5.81GSLQDD262 pKa = 3.44DD263 pKa = 4.17CKK265 pKa = 10.62SARR268 pKa = 11.84VPITVTVNAKK278 pKa = 8.69PTIPVAPGNIQNICPDD294 pKa = 3.41TTADD298 pKa = 3.7LTTIDD303 pKa = 4.61LTASTPGGFFEE314 pKa = 4.52WRR316 pKa = 11.84EE317 pKa = 4.05GMLSTSALVDD327 pKa = 3.84DD328 pKa = 4.35PTKK331 pKa = 11.0VEE333 pKa = 3.81AGTYY337 pKa = 7.16YY338 pKa = 9.83ICEE341 pKa = 4.17KK342 pKa = 10.65SVEE345 pKa = 4.11GCYY348 pKa = 10.71GSAQAVVVNIVPCDD362 pKa = 3.64CQLEE366 pKa = 4.26YY367 pKa = 11.09SVAAGVDD374 pKa = 3.58QEE376 pKa = 4.65VCAGMPISVTATTSGTVSGIIWTTSGTGTFANADD410 pKa = 4.1SLNTTYY416 pKa = 10.39TPSAADD422 pKa = 3.16IAAGEE427 pKa = 4.18IALTVTTNDD436 pKa = 3.42PDD438 pKa = 5.37GDD440 pKa = 4.09DD441 pKa = 3.65TCVPKK446 pKa = 10.03MDD448 pKa = 4.67ALTVVIIPQPAPAYY462 pKa = 10.08GVACDD467 pKa = 4.17DD468 pKa = 3.87TLICVGKK475 pKa = 8.25STKK478 pKa = 10.6LLGFAPGYY486 pKa = 7.85TINWYY491 pKa = 5.8TTPTGGTPIGTTPSGGKK508 pKa = 8.31LTVSPSATTTYY519 pKa = 9.65YY520 pKa = 11.23AEE522 pKa = 5.05AIGDD526 pKa = 3.86KK527 pKa = 10.51GCSSEE532 pKa = 3.92EE533 pKa = 3.91RR534 pKa = 11.84TPVTVVVQPCSSDD547 pKa = 3.27LAIVKK552 pKa = 8.46TVLTPAPYY560 pKa = 10.25SAGQEE565 pKa = 3.86INYY568 pKa = 10.03SLTVTSLAVGNASNVTVEE586 pKa = 4.43DD587 pKa = 3.77VLPASLLYY595 pKa = 10.44VASAPAGEE603 pKa = 4.52YY604 pKa = 9.86NAEE607 pKa = 4.13TGVWTIGNMINGSNRR622 pKa = 11.84VLVITAKK629 pKa = 10.39IQDD632 pKa = 3.99NASGDD637 pKa = 3.53ITNTAIVKK645 pKa = 10.64SPDD648 pKa = 3.34NDD650 pKa = 3.93PGNTSNDD657 pKa = 3.45TSSVTIRR664 pKa = 11.84VGNLADD670 pKa = 4.32LMLAKK675 pKa = 10.09KK676 pKa = 10.56VSTVNPALGEE686 pKa = 4.47SITYY690 pKa = 8.36TLEE693 pKa = 3.7VSNNGPQRR701 pKa = 11.84ATNIEE706 pKa = 4.1VTDD709 pKa = 3.67QLPAGLEE716 pKa = 4.45FVSSTDD722 pKa = 3.72FSKK725 pKa = 9.8TGNLLKK731 pKa = 10.32GTIDD735 pKa = 3.76SIEE738 pKa = 4.14VGDD741 pKa = 4.05TKK743 pKa = 10.93TLTFLAKK750 pKa = 9.95VVSGTSITNIAQISKK765 pKa = 10.31SDD767 pKa = 3.49QKK769 pKa = 11.61DD770 pKa = 3.41PDD772 pKa = 3.82STPGNGYY779 pKa = 10.97SNGEE783 pKa = 4.0DD784 pKa = 3.81DD785 pKa = 4.25EE786 pKa = 6.63ASVTVNTGCPTIDD799 pKa = 3.79PPIIACAQTTICVGTSVTLTAVGCKK824 pKa = 10.04NGTVKK829 pKa = 10.32WSNGMEE835 pKa = 4.18GVSITLPIDD844 pKa = 3.11QTTTFTAVCQKK855 pKa = 10.64DD856 pKa = 3.67EE857 pKa = 4.59CNSIPSNPITINVANTVKK875 pKa = 10.35PVLASNVSSVCEE887 pKa = 4.21GGSATLTAANCNGVLVWSTGATGTPWW913 pKa = 3.32
MM1 pKa = 7.41VDD3 pKa = 3.38GVTVAVVNSDD13 pKa = 3.14STLTIKK19 pKa = 11.01GPGSYY24 pKa = 9.09TFLSTSGVLYY34 pKa = 9.42PASGFCAIEE43 pKa = 4.07VVPGAFFDD51 pKa = 4.22LALSKK56 pKa = 10.9SLAPGQASTVTAGSLVKK73 pKa = 10.28FRR75 pKa = 11.84LTVSNEE81 pKa = 3.59GNMEE85 pKa = 4.24ALQIALSDD93 pKa = 3.86SLPEE97 pKa = 4.23GMTLADD103 pKa = 4.02TSWTASGGFATLNVPLAGPLYY124 pKa = 9.86PGATAFIDD132 pKa = 3.03ITLQVDD138 pKa = 3.42SSFTSGTLTNFAQIKK153 pKa = 9.52DD154 pKa = 3.77ASCATDD160 pKa = 3.78EE161 pKa = 4.29PVIDD165 pKa = 4.86IDD167 pKa = 3.92STPDD171 pKa = 2.93NGFNNGEE178 pKa = 4.46DD179 pKa = 4.13DD180 pKa = 5.27DD181 pKa = 5.54DD182 pKa = 5.46SEE184 pKa = 5.41SISISQCPTITLTTSSDD201 pKa = 3.2KK202 pKa = 11.03VICEE206 pKa = 4.23GEE208 pKa = 4.45SVVLNVTASEE218 pKa = 4.21SGATIDD224 pKa = 3.79WYY226 pKa = 9.52TVPTGGTPFVTTASEE241 pKa = 4.12ADD243 pKa = 3.47QTLSPTQTTTYY254 pKa = 9.61YY255 pKa = 10.92VEE257 pKa = 5.81GSLQDD262 pKa = 3.44DD263 pKa = 4.17CKK265 pKa = 10.62SARR268 pKa = 11.84VPITVTVNAKK278 pKa = 8.69PTIPVAPGNIQNICPDD294 pKa = 3.41TTADD298 pKa = 3.7LTTIDD303 pKa = 4.61LTASTPGGFFEE314 pKa = 4.52WRR316 pKa = 11.84EE317 pKa = 4.05GMLSTSALVDD327 pKa = 3.84DD328 pKa = 4.35PTKK331 pKa = 11.0VEE333 pKa = 3.81AGTYY337 pKa = 7.16YY338 pKa = 9.83ICEE341 pKa = 4.17KK342 pKa = 10.65SVEE345 pKa = 4.11GCYY348 pKa = 10.71GSAQAVVVNIVPCDD362 pKa = 3.64CQLEE366 pKa = 4.26YY367 pKa = 11.09SVAAGVDD374 pKa = 3.58QEE376 pKa = 4.65VCAGMPISVTATTSGTVSGIIWTTSGTGTFANADD410 pKa = 4.1SLNTTYY416 pKa = 10.39TPSAADD422 pKa = 3.16IAAGEE427 pKa = 4.18IALTVTTNDD436 pKa = 3.42PDD438 pKa = 5.37GDD440 pKa = 4.09DD441 pKa = 3.65TCVPKK446 pKa = 10.03MDD448 pKa = 4.67ALTVVIIPQPAPAYY462 pKa = 10.08GVACDD467 pKa = 4.17DD468 pKa = 3.87TLICVGKK475 pKa = 8.25STKK478 pKa = 10.6LLGFAPGYY486 pKa = 7.85TINWYY491 pKa = 5.8TTPTGGTPIGTTPSGGKK508 pKa = 8.31LTVSPSATTTYY519 pKa = 9.65YY520 pKa = 11.23AEE522 pKa = 5.05AIGDD526 pKa = 3.86KK527 pKa = 10.51GCSSEE532 pKa = 3.92EE533 pKa = 3.91RR534 pKa = 11.84TPVTVVVQPCSSDD547 pKa = 3.27LAIVKK552 pKa = 8.46TVLTPAPYY560 pKa = 10.25SAGQEE565 pKa = 3.86INYY568 pKa = 10.03SLTVTSLAVGNASNVTVEE586 pKa = 4.43DD587 pKa = 3.77VLPASLLYY595 pKa = 10.44VASAPAGEE603 pKa = 4.52YY604 pKa = 9.86NAEE607 pKa = 4.13TGVWTIGNMINGSNRR622 pKa = 11.84VLVITAKK629 pKa = 10.39IQDD632 pKa = 3.99NASGDD637 pKa = 3.53ITNTAIVKK645 pKa = 10.64SPDD648 pKa = 3.34NDD650 pKa = 3.93PGNTSNDD657 pKa = 3.45TSSVTIRR664 pKa = 11.84VGNLADD670 pKa = 4.32LMLAKK675 pKa = 10.09KK676 pKa = 10.56VSTVNPALGEE686 pKa = 4.47SITYY690 pKa = 8.36TLEE693 pKa = 3.7VSNNGPQRR701 pKa = 11.84ATNIEE706 pKa = 4.1VTDD709 pKa = 3.67QLPAGLEE716 pKa = 4.45FVSSTDD722 pKa = 3.72FSKK725 pKa = 9.8TGNLLKK731 pKa = 10.32GTIDD735 pKa = 3.76SIEE738 pKa = 4.14VGDD741 pKa = 4.05TKK743 pKa = 10.93TLTFLAKK750 pKa = 9.95VVSGTSITNIAQISKK765 pKa = 10.31SDD767 pKa = 3.49QKK769 pKa = 11.61DD770 pKa = 3.41PDD772 pKa = 3.82STPGNGYY779 pKa = 10.97SNGEE783 pKa = 4.0DD784 pKa = 3.81DD785 pKa = 4.25EE786 pKa = 6.63ASVTVNTGCPTIDD799 pKa = 3.79PPIIACAQTTICVGTSVTLTAVGCKK824 pKa = 10.04NGTVKK829 pKa = 10.32WSNGMEE835 pKa = 4.18GVSITLPIDD844 pKa = 3.11QTTTFTAVCQKK855 pKa = 10.64DD856 pKa = 3.67EE857 pKa = 4.59CNSIPSNPITINVANTVKK875 pKa = 10.35PVLASNVSSVCEE887 pKa = 4.21GGSATLTAANCNGVLVWSTGATGTPWW913 pKa = 3.32
Molecular weight: 93.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C9BLC4|A0A7C9BLC4_9BACT Uncharacterized protein OS=Cytophagaceae bacterium SJW1-29 OX=2654236 GN=GBK04_26170 PE=4 SV=1
MM1 pKa = 7.84LITHH5 pKa = 6.35TTDD8 pKa = 3.34QQVCCPLCQQVSSRR22 pKa = 11.84LHH24 pKa = 4.89GFYY27 pKa = 10.34YY28 pKa = 10.49RR29 pKa = 11.84KK30 pKa = 8.74PTDD33 pKa = 3.68LFIGDD38 pKa = 3.94KK39 pKa = 10.22QVQLRR44 pKa = 11.84VRR46 pKa = 11.84LRR48 pKa = 11.84RR49 pKa = 11.84FRR51 pKa = 11.84CLNSVRR57 pKa = 11.84PKK59 pKa = 9.76RR60 pKa = 11.84TFGQPCPDD68 pKa = 2.91WLFTFFHH75 pKa = 6.06RR76 pKa = 11.84TSLLVHH82 pKa = 6.33AQRR85 pKa = 11.84NVAMVLGGKK94 pKa = 8.25TSARR98 pKa = 11.84LLIHH102 pKa = 6.51LHH104 pKa = 5.9MLTSHH109 pKa = 6.81DD110 pKa = 3.55TLVRR114 pKa = 11.84IIRR117 pKa = 11.84KK118 pKa = 5.41WQPVDD123 pKa = 3.46LQTPHH128 pKa = 7.13ALSVDD133 pKa = 2.91DD134 pKa = 3.74WAIRR138 pKa = 11.84KK139 pKa = 8.97AKK141 pKa = 9.84TYY143 pKa = 8.97GTILTVRR150 pKa = 11.84RR151 pKa = 11.84YY152 pKa = 9.91DD153 pKa = 3.18
MM1 pKa = 7.84LITHH5 pKa = 6.35TTDD8 pKa = 3.34QQVCCPLCQQVSSRR22 pKa = 11.84LHH24 pKa = 4.89GFYY27 pKa = 10.34YY28 pKa = 10.49RR29 pKa = 11.84KK30 pKa = 8.74PTDD33 pKa = 3.68LFIGDD38 pKa = 3.94KK39 pKa = 10.22QVQLRR44 pKa = 11.84VRR46 pKa = 11.84LRR48 pKa = 11.84RR49 pKa = 11.84FRR51 pKa = 11.84CLNSVRR57 pKa = 11.84PKK59 pKa = 9.76RR60 pKa = 11.84TFGQPCPDD68 pKa = 2.91WLFTFFHH75 pKa = 6.06RR76 pKa = 11.84TSLLVHH82 pKa = 6.33AQRR85 pKa = 11.84NVAMVLGGKK94 pKa = 8.25TSARR98 pKa = 11.84LLIHH102 pKa = 6.51LHH104 pKa = 5.9MLTSHH109 pKa = 6.81DD110 pKa = 3.55TLVRR114 pKa = 11.84IIRR117 pKa = 11.84KK118 pKa = 5.41WQPVDD123 pKa = 3.46LQTPHH128 pKa = 7.13ALSVDD133 pKa = 2.91DD134 pKa = 3.74WAIRR138 pKa = 11.84KK139 pKa = 8.97AKK141 pKa = 9.84TYY143 pKa = 8.97GTILTVRR150 pKa = 11.84RR151 pKa = 11.84YY152 pKa = 9.91DD153 pKa = 3.18
Molecular weight: 17.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1951742 |
20 |
2550 |
346.7 |
38.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.848 ± 0.034 | 0.751 ± 0.011 |
5.388 ± 0.023 | 5.921 ± 0.034 |
4.652 ± 0.026 | 7.258 ± 0.034 |
1.888 ± 0.018 | 5.951 ± 0.027 |
5.555 ± 0.03 | 9.986 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.013 | 4.646 ± 0.028 |
4.485 ± 0.022 | 3.99 ± 0.02 |
5.164 ± 0.027 | 6.175 ± 0.025 |
6.041 ± 0.032 | 6.685 ± 0.025 |
1.381 ± 0.014 | 4.0 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
