
Junonia coenia densovirus (isolate pBRJ/1990) (JcDNV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Densovirinae; Ambidensovirus; Lepidopteran ambidensovirus 1; Junonia coenia densovirus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
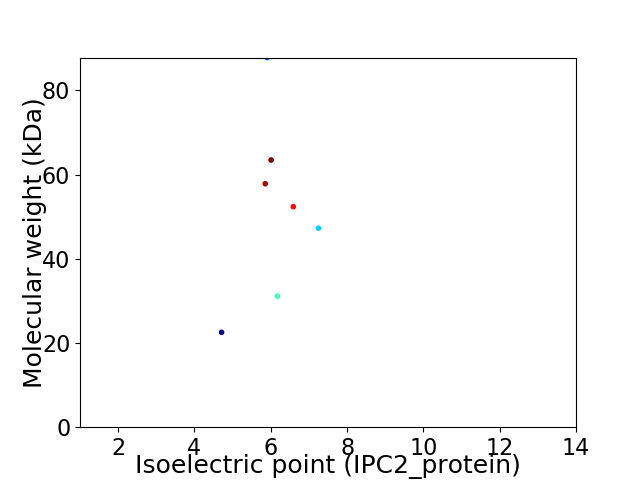
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
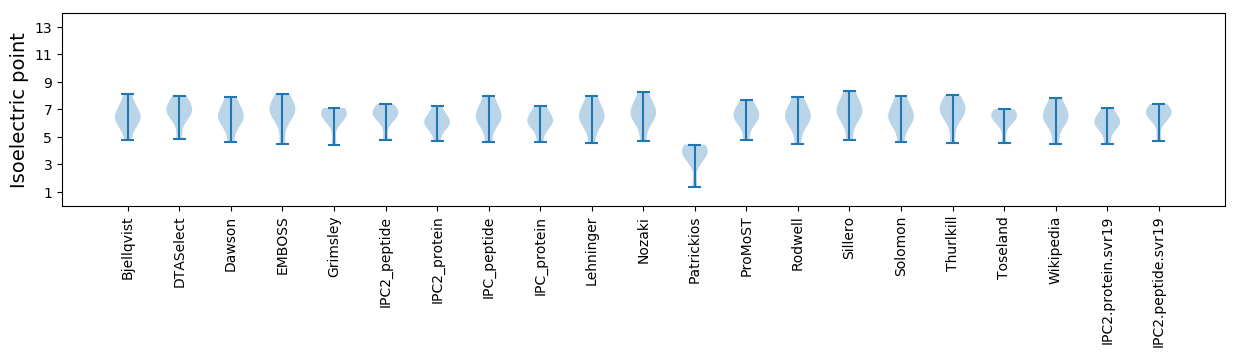
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q90053-2|CAPSD-2_JDNVP Isoform of Q90053 Isoform VP2 of Capsid protein VP1 OS=Junonia coenia densovirus (isolate pBRJ/1990) OX=648250 GN=VP PE=4 SV=1
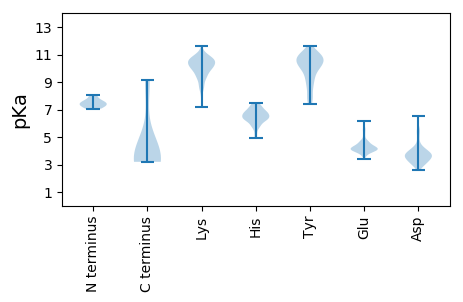
MM1 pKa = 7.22QHH3 pKa = 7.19DD4 pKa = 5.37LDD6 pKa = 4.12NQIIACGEE14 pKa = 4.68DD15 pKa = 3.31IDD17 pKa = 4.49HH18 pKa = 6.43TLAMEE23 pKa = 4.49EE24 pKa = 4.49LEE26 pKa = 4.07HH27 pKa = 7.24WDD29 pKa = 2.94WTKK32 pKa = 11.08QNRR35 pKa = 11.84LPFQLYY41 pKa = 9.49LAVMHH46 pKa = 6.64LNEE49 pKa = 4.17IPEE52 pKa = 4.24WLDD55 pKa = 2.89EE56 pKa = 4.26TMLIEE61 pKa = 3.85CVYY64 pKa = 10.39YY65 pKa = 10.45FKK67 pKa = 11.1EE68 pKa = 4.67LINHH72 pKa = 6.96RR73 pKa = 11.84DD74 pKa = 3.5PYY76 pKa = 10.9DD77 pKa = 3.56TDD79 pKa = 4.78EE80 pKa = 4.42FNAWNMNGKK89 pKa = 8.73PFKK92 pKa = 10.53TMWKK96 pKa = 8.76ICKK99 pKa = 9.35FCYY102 pKa = 9.5TNCEE106 pKa = 4.11DD107 pKa = 3.9PDD109 pKa = 3.92EE110 pKa = 4.09YY111 pKa = 11.39RR112 pKa = 11.84FMYY115 pKa = 10.02NRR117 pKa = 11.84TVFVEE122 pKa = 4.08DD123 pKa = 4.35AEE125 pKa = 5.28DD126 pKa = 3.96IINRR130 pKa = 11.84LQDD133 pKa = 3.38GSSWCQICHH142 pKa = 6.19TCPLFNISVIYY153 pKa = 10.32EE154 pKa = 3.68NSPNKK159 pKa = 9.24KK160 pKa = 9.17RR161 pKa = 11.84RR162 pKa = 11.84YY163 pKa = 8.71SSSSEE168 pKa = 3.39EE169 pKa = 4.04DD170 pKa = 3.19EE171 pKa = 4.36YY172 pKa = 11.21MSNSFYY178 pKa = 11.24VKK180 pKa = 10.26HH181 pKa = 6.3PNSRR185 pKa = 11.84HH186 pKa = 4.18
MM1 pKa = 7.22QHH3 pKa = 7.19DD4 pKa = 5.37LDD6 pKa = 4.12NQIIACGEE14 pKa = 4.68DD15 pKa = 3.31IDD17 pKa = 4.49HH18 pKa = 6.43TLAMEE23 pKa = 4.49EE24 pKa = 4.49LEE26 pKa = 4.07HH27 pKa = 7.24WDD29 pKa = 2.94WTKK32 pKa = 11.08QNRR35 pKa = 11.84LPFQLYY41 pKa = 9.49LAVMHH46 pKa = 6.64LNEE49 pKa = 4.17IPEE52 pKa = 4.24WLDD55 pKa = 2.89EE56 pKa = 4.26TMLIEE61 pKa = 3.85CVYY64 pKa = 10.39YY65 pKa = 10.45FKK67 pKa = 11.1EE68 pKa = 4.67LINHH72 pKa = 6.96RR73 pKa = 11.84DD74 pKa = 3.5PYY76 pKa = 10.9DD77 pKa = 3.56TDD79 pKa = 4.78EE80 pKa = 4.42FNAWNMNGKK89 pKa = 8.73PFKK92 pKa = 10.53TMWKK96 pKa = 8.76ICKK99 pKa = 9.35FCYY102 pKa = 9.5TNCEE106 pKa = 4.11DD107 pKa = 3.9PDD109 pKa = 3.92EE110 pKa = 4.09YY111 pKa = 11.39RR112 pKa = 11.84FMYY115 pKa = 10.02NRR117 pKa = 11.84TVFVEE122 pKa = 4.08DD123 pKa = 4.35AEE125 pKa = 5.28DD126 pKa = 3.96IINRR130 pKa = 11.84LQDD133 pKa = 3.38GSSWCQICHH142 pKa = 6.19TCPLFNISVIYY153 pKa = 10.32EE154 pKa = 3.68NSPNKK159 pKa = 9.24KK160 pKa = 9.17RR161 pKa = 11.84RR162 pKa = 11.84YY163 pKa = 8.71SSSSEE168 pKa = 3.39EE169 pKa = 4.04DD170 pKa = 3.19EE171 pKa = 4.36YY172 pKa = 11.21MSNSFYY178 pKa = 11.24VKK180 pKa = 10.26HH181 pKa = 6.3PNSRR185 pKa = 11.84HH186 pKa = 4.18
Molecular weight: 22.54 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q90053-4|CAPSD-4_JDNVP Isoform of Q90053 Isoform VP4 of Capsid protein VP1 OS=Junonia coenia densovirus (isolate pBRJ/1990) OX=648250 GN=VP PE=4 SV=1
MM1 pKa = 7.58SLPGTGSGTSSGGGNTSGQEE21 pKa = 4.0VYY23 pKa = 10.71VIPRR27 pKa = 11.84PFSNFGKK34 pKa = 10.62KK35 pKa = 10.08LSTYY39 pKa = 7.81TKK41 pKa = 8.62SHH43 pKa = 6.28KK44 pKa = 10.63FMIFGLANNVIGPTGTGTTAVNRR67 pKa = 11.84LITTCLAEE75 pKa = 4.61IPWQKK80 pKa = 10.89LPLYY84 pKa = 8.6MNQSEE89 pKa = 4.69FDD91 pKa = 4.02LLPPGSRR98 pKa = 11.84VVEE101 pKa = 4.39CNVKK105 pKa = 10.69VIFRR109 pKa = 11.84TNRR112 pKa = 11.84IAFEE116 pKa = 4.15TSSTATKK123 pKa = 10.31QATLNQISNLQTAVGLNKK141 pKa = 10.33LGWGIDD147 pKa = 3.23RR148 pKa = 11.84SFTAFQSDD156 pKa = 3.51QPMIPTATSAPKK168 pKa = 10.21YY169 pKa = 10.12EE170 pKa = 5.64PITGTTGYY178 pKa = 10.42RR179 pKa = 11.84GMIADD184 pKa = 4.09YY185 pKa = 11.0YY186 pKa = 11.09GADD189 pKa = 3.73STNDD193 pKa = 3.38AAFGNAGNYY202 pKa = 7.38PHH204 pKa = 6.82HH205 pKa = 6.4QVGSFTFIQNYY216 pKa = 7.86YY217 pKa = 11.13CMYY220 pKa = 9.91QQTNQGTGGWPCLAEE235 pKa = 4.26HH236 pKa = 6.78LQQFDD241 pKa = 3.76SKK243 pKa = 9.86TVNNQCLIDD252 pKa = 3.78VTYY255 pKa = 9.34KK256 pKa = 10.87PKK258 pKa = 10.0MGLIKK263 pKa = 10.54PPLNYY268 pKa = 10.1KK269 pKa = 10.37IIGQPTAKK277 pKa = 9.41GTISVGDD284 pKa = 3.66NLVNMRR290 pKa = 11.84GAVVINPPEE299 pKa = 4.13ATQSVTEE306 pKa = 4.27STHH309 pKa = 5.8NLTRR313 pKa = 11.84NFPANLFNIYY323 pKa = 10.6SDD325 pKa = 3.59IEE327 pKa = 4.02KK328 pKa = 10.69SQILHH333 pKa = 6.25KK334 pKa = 10.78GPWGHH339 pKa = 6.63EE340 pKa = 3.99NPQIQPSVHH349 pKa = 7.13IGIQAVPALTTGALLVNSSPLNSWTDD375 pKa = 3.1SMGYY379 pKa = 8.99IDD381 pKa = 5.55VMSSCTVMEE390 pKa = 4.21SQPTHH395 pKa = 6.45FPFSTDD401 pKa = 2.82ANTNPGNTIYY411 pKa = 10.77RR412 pKa = 11.84INLTPNSLTSAFNGLYY428 pKa = 10.89GNGATLGNVV437 pKa = 3.2
MM1 pKa = 7.58SLPGTGSGTSSGGGNTSGQEE21 pKa = 4.0VYY23 pKa = 10.71VIPRR27 pKa = 11.84PFSNFGKK34 pKa = 10.62KK35 pKa = 10.08LSTYY39 pKa = 7.81TKK41 pKa = 8.62SHH43 pKa = 6.28KK44 pKa = 10.63FMIFGLANNVIGPTGTGTTAVNRR67 pKa = 11.84LITTCLAEE75 pKa = 4.61IPWQKK80 pKa = 10.89LPLYY84 pKa = 8.6MNQSEE89 pKa = 4.69FDD91 pKa = 4.02LLPPGSRR98 pKa = 11.84VVEE101 pKa = 4.39CNVKK105 pKa = 10.69VIFRR109 pKa = 11.84TNRR112 pKa = 11.84IAFEE116 pKa = 4.15TSSTATKK123 pKa = 10.31QATLNQISNLQTAVGLNKK141 pKa = 10.33LGWGIDD147 pKa = 3.23RR148 pKa = 11.84SFTAFQSDD156 pKa = 3.51QPMIPTATSAPKK168 pKa = 10.21YY169 pKa = 10.12EE170 pKa = 5.64PITGTTGYY178 pKa = 10.42RR179 pKa = 11.84GMIADD184 pKa = 4.09YY185 pKa = 11.0YY186 pKa = 11.09GADD189 pKa = 3.73STNDD193 pKa = 3.38AAFGNAGNYY202 pKa = 7.38PHH204 pKa = 6.82HH205 pKa = 6.4QVGSFTFIQNYY216 pKa = 7.86YY217 pKa = 11.13CMYY220 pKa = 9.91QQTNQGTGGWPCLAEE235 pKa = 4.26HH236 pKa = 6.78LQQFDD241 pKa = 3.76SKK243 pKa = 9.86TVNNQCLIDD252 pKa = 3.78VTYY255 pKa = 9.34KK256 pKa = 10.87PKK258 pKa = 10.0MGLIKK263 pKa = 10.54PPLNYY268 pKa = 10.1KK269 pKa = 10.37IIGQPTAKK277 pKa = 9.41GTISVGDD284 pKa = 3.66NLVNMRR290 pKa = 11.84GAVVINPPEE299 pKa = 4.13ATQSVTEE306 pKa = 4.27STHH309 pKa = 5.8NLTRR313 pKa = 11.84NFPANLFNIYY323 pKa = 10.6SDD325 pKa = 3.59IEE327 pKa = 4.02KK328 pKa = 10.69SQILHH333 pKa = 6.25KK334 pKa = 10.78GPWGHH339 pKa = 6.63EE340 pKa = 3.99NPQIQPSVHH349 pKa = 7.13IGIQAVPALTTGALLVNSSPLNSWTDD375 pKa = 3.1SMGYY379 pKa = 8.99IDD381 pKa = 5.55VMSSCTVMEE390 pKa = 4.21SQPTHH395 pKa = 6.45FPFSTDD401 pKa = 2.82ANTNPGNTIYY411 pKa = 10.77RR412 pKa = 11.84INLTPNSLTSAFNGLYY428 pKa = 10.89GNGATLGNVV437 pKa = 3.2
Molecular weight: 47.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3274 |
186 |
810 |
467.7 |
51.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.864 ± 0.543 | 1.374 ± 0.292 |
4.368 ± 0.56 | 5.192 ± 0.748 |
3.971 ± 0.419 | 7.697 ± 0.95 |
2.23 ± 0.182 | 5.987 ± 0.156 |
5.192 ± 0.487 | 6.964 ± 0.303 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.535 ± 0.228 | 7.575 ± 0.353 |
5.834 ± 0.497 | 4.673 ± 0.309 |
3.451 ± 0.654 | 8.308 ± 0.418 |
8.369 ± 0.813 | 4.918 ± 0.227 |
1.374 ± 0.265 | 4.032 ± 0.23 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
