
Human herpesvirus 1 (strain 17) (HHV-1) (Human herpes simplex virus 1)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Simplexvirus; Human alphaherpesvirus 1
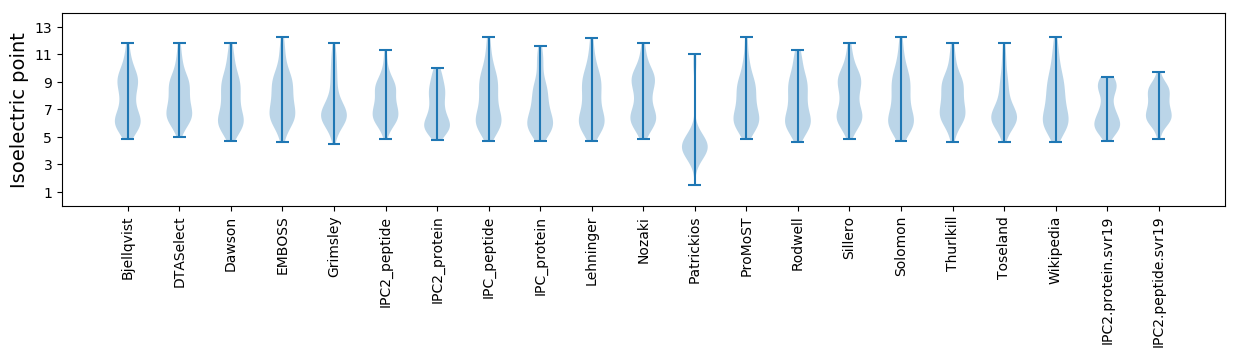
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 77 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
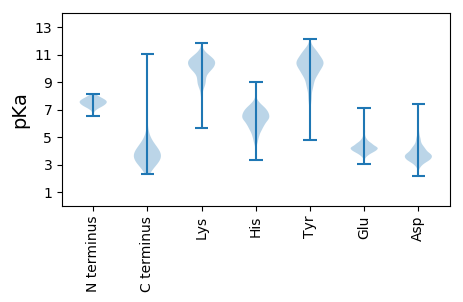
Protein with the lowest isoelectric point:
>sp|P08392|ICP4_HHV11 Major viral transcription factor ICP4 OS=Human herpesvirus 1 (strain 17) OX=10299 GN=ICP4 PE=1 SV=1
MM1 pKa = 7.97DD2 pKa = 5.67LLVDD6 pKa = 4.15EE7 pKa = 5.76LFADD11 pKa = 4.29MNADD15 pKa = 3.63GASPPPPRR23 pKa = 11.84PAGGPKK29 pKa = 8.47NTPAAPPLYY38 pKa = 9.66ATGRR42 pKa = 11.84LSQAQLMPSPPMPVPPAALFNRR64 pKa = 11.84LLDD67 pKa = 4.24DD68 pKa = 4.82LGFSAGPALCTMLDD82 pKa = 3.31TWNEE86 pKa = 4.08DD87 pKa = 3.39LFSALPTNADD97 pKa = 4.16LYY99 pKa = 10.72RR100 pKa = 11.84EE101 pKa = 4.37CKK103 pKa = 9.8FLSTLPSDD111 pKa = 3.77VVEE114 pKa = 4.41WGDD117 pKa = 3.38AYY119 pKa = 11.29VPEE122 pKa = 4.45RR123 pKa = 11.84TQIDD127 pKa = 3.18IRR129 pKa = 11.84AHH131 pKa = 5.99GDD133 pKa = 3.29VAFPTLPATRR143 pKa = 11.84DD144 pKa = 3.59GLGLYY149 pKa = 10.03YY150 pKa = 10.38EE151 pKa = 4.24ALSRR155 pKa = 11.84FFHH158 pKa = 6.56AEE160 pKa = 3.02LRR162 pKa = 11.84ARR164 pKa = 11.84EE165 pKa = 3.89EE166 pKa = 4.02SYY168 pKa = 9.67RR169 pKa = 11.84TVLANFCSALYY180 pKa = 9.82RR181 pKa = 11.84YY182 pKa = 9.56LRR184 pKa = 11.84ASVRR188 pKa = 11.84QLHH191 pKa = 5.87RR192 pKa = 11.84QAHH195 pKa = 4.64MRR197 pKa = 11.84GRR199 pKa = 11.84DD200 pKa = 3.25RR201 pKa = 11.84DD202 pKa = 3.74LGEE205 pKa = 4.07MLRR208 pKa = 11.84ATIADD213 pKa = 3.86RR214 pKa = 11.84YY215 pKa = 8.55YY216 pKa = 11.06RR217 pKa = 11.84EE218 pKa = 3.99TARR221 pKa = 11.84LARR224 pKa = 11.84VLFLHH229 pKa = 7.13LYY231 pKa = 10.43LFLTRR236 pKa = 11.84EE237 pKa = 4.38ILWAAYY243 pKa = 9.3AEE245 pKa = 4.24QMMRR249 pKa = 11.84PDD251 pKa = 6.0LFDD254 pKa = 4.82CLCCDD259 pKa = 4.03LEE261 pKa = 4.33SWRR264 pKa = 11.84QLAGLFQPFMFVNGALTVRR283 pKa = 11.84GVPIEE288 pKa = 3.76ARR290 pKa = 11.84RR291 pKa = 11.84LRR293 pKa = 11.84EE294 pKa = 3.74LNHH297 pKa = 6.26IRR299 pKa = 11.84EE300 pKa = 4.48HH301 pKa = 6.62LNLPLVRR308 pKa = 11.84SAATEE313 pKa = 4.05EE314 pKa = 4.29PGAPLTTPPTLHH326 pKa = 6.67GNQARR331 pKa = 11.84ASGYY335 pKa = 10.28FMVLIRR341 pKa = 11.84AKK343 pKa = 10.19LDD345 pKa = 3.45SYY347 pKa = 11.91SSFTTSPSEE356 pKa = 3.72AVMRR360 pKa = 11.84EE361 pKa = 3.85HH362 pKa = 7.26AYY364 pKa = 10.74SRR366 pKa = 11.84ARR368 pKa = 11.84TKK370 pKa = 11.08NNYY373 pKa = 9.6GSTIEE378 pKa = 4.57GLLDD382 pKa = 5.03LPDD385 pKa = 4.99DD386 pKa = 4.89DD387 pKa = 5.62APEE390 pKa = 4.23EE391 pKa = 4.33AGLAAPRR398 pKa = 11.84LSFLPAGHH406 pKa = 6.02TRR408 pKa = 11.84RR409 pKa = 11.84LSTAPPTDD417 pKa = 3.06VSLGDD422 pKa = 3.75EE423 pKa = 4.16LHH425 pKa = 7.34LDD427 pKa = 4.01GEE429 pKa = 4.85DD430 pKa = 3.13VAMAHH435 pKa = 7.2ADD437 pKa = 3.53ALDD440 pKa = 4.82DD441 pKa = 4.82FDD443 pKa = 7.35LDD445 pKa = 3.79MLGDD449 pKa = 4.16GDD451 pKa = 4.27SPGPGFTPHH460 pKa = 7.46DD461 pKa = 4.07SAPYY465 pKa = 9.65GALDD469 pKa = 3.39MADD472 pKa = 4.37FEE474 pKa = 4.54FEE476 pKa = 4.81QMFTDD481 pKa = 3.78ALGIDD486 pKa = 4.23EE487 pKa = 4.57YY488 pKa = 11.73GGG490 pKa = 3.43
MM1 pKa = 7.97DD2 pKa = 5.67LLVDD6 pKa = 4.15EE7 pKa = 5.76LFADD11 pKa = 4.29MNADD15 pKa = 3.63GASPPPPRR23 pKa = 11.84PAGGPKK29 pKa = 8.47NTPAAPPLYY38 pKa = 9.66ATGRR42 pKa = 11.84LSQAQLMPSPPMPVPPAALFNRR64 pKa = 11.84LLDD67 pKa = 4.24DD68 pKa = 4.82LGFSAGPALCTMLDD82 pKa = 3.31TWNEE86 pKa = 4.08DD87 pKa = 3.39LFSALPTNADD97 pKa = 4.16LYY99 pKa = 10.72RR100 pKa = 11.84EE101 pKa = 4.37CKK103 pKa = 9.8FLSTLPSDD111 pKa = 3.77VVEE114 pKa = 4.41WGDD117 pKa = 3.38AYY119 pKa = 11.29VPEE122 pKa = 4.45RR123 pKa = 11.84TQIDD127 pKa = 3.18IRR129 pKa = 11.84AHH131 pKa = 5.99GDD133 pKa = 3.29VAFPTLPATRR143 pKa = 11.84DD144 pKa = 3.59GLGLYY149 pKa = 10.03YY150 pKa = 10.38EE151 pKa = 4.24ALSRR155 pKa = 11.84FFHH158 pKa = 6.56AEE160 pKa = 3.02LRR162 pKa = 11.84ARR164 pKa = 11.84EE165 pKa = 3.89EE166 pKa = 4.02SYY168 pKa = 9.67RR169 pKa = 11.84TVLANFCSALYY180 pKa = 9.82RR181 pKa = 11.84YY182 pKa = 9.56LRR184 pKa = 11.84ASVRR188 pKa = 11.84QLHH191 pKa = 5.87RR192 pKa = 11.84QAHH195 pKa = 4.64MRR197 pKa = 11.84GRR199 pKa = 11.84DD200 pKa = 3.25RR201 pKa = 11.84DD202 pKa = 3.74LGEE205 pKa = 4.07MLRR208 pKa = 11.84ATIADD213 pKa = 3.86RR214 pKa = 11.84YY215 pKa = 8.55YY216 pKa = 11.06RR217 pKa = 11.84EE218 pKa = 3.99TARR221 pKa = 11.84LARR224 pKa = 11.84VLFLHH229 pKa = 7.13LYY231 pKa = 10.43LFLTRR236 pKa = 11.84EE237 pKa = 4.38ILWAAYY243 pKa = 9.3AEE245 pKa = 4.24QMMRR249 pKa = 11.84PDD251 pKa = 6.0LFDD254 pKa = 4.82CLCCDD259 pKa = 4.03LEE261 pKa = 4.33SWRR264 pKa = 11.84QLAGLFQPFMFVNGALTVRR283 pKa = 11.84GVPIEE288 pKa = 3.76ARR290 pKa = 11.84RR291 pKa = 11.84LRR293 pKa = 11.84EE294 pKa = 3.74LNHH297 pKa = 6.26IRR299 pKa = 11.84EE300 pKa = 4.48HH301 pKa = 6.62LNLPLVRR308 pKa = 11.84SAATEE313 pKa = 4.05EE314 pKa = 4.29PGAPLTTPPTLHH326 pKa = 6.67GNQARR331 pKa = 11.84ASGYY335 pKa = 10.28FMVLIRR341 pKa = 11.84AKK343 pKa = 10.19LDD345 pKa = 3.45SYY347 pKa = 11.91SSFTTSPSEE356 pKa = 3.72AVMRR360 pKa = 11.84EE361 pKa = 3.85HH362 pKa = 7.26AYY364 pKa = 10.74SRR366 pKa = 11.84ARR368 pKa = 11.84TKK370 pKa = 11.08NNYY373 pKa = 9.6GSTIEE378 pKa = 4.57GLLDD382 pKa = 5.03LPDD385 pKa = 4.99DD386 pKa = 4.89DD387 pKa = 5.62APEE390 pKa = 4.23EE391 pKa = 4.33AGLAAPRR398 pKa = 11.84LSFLPAGHH406 pKa = 6.02TRR408 pKa = 11.84RR409 pKa = 11.84LSTAPPTDD417 pKa = 3.06VSLGDD422 pKa = 3.75EE423 pKa = 4.16LHH425 pKa = 7.34LDD427 pKa = 4.01GEE429 pKa = 4.85DD430 pKa = 3.13VAMAHH435 pKa = 7.2ADD437 pKa = 3.53ALDD440 pKa = 4.82DD441 pKa = 4.82FDD443 pKa = 7.35LDD445 pKa = 3.79MLGDD449 pKa = 4.16GDD451 pKa = 4.27SPGPGFTPHH460 pKa = 7.46DD461 pKa = 4.07SAPYY465 pKa = 9.65GALDD469 pKa = 3.39MADD472 pKa = 4.37FEE474 pKa = 4.54FEE476 pKa = 4.81QMFTDD481 pKa = 3.78ALGIDD486 pKa = 4.23EE487 pKa = 4.57YY488 pKa = 11.73GGG490 pKa = 3.43
Molecular weight: 54.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P10234|DUT_HHV11 Deoxyuridine 5'-triphosphate nucleotidohydrolase OS=Human herpesvirus 1 (strain 17) OX=10299 GN=DUT PE=3 SV=1
MM1 pKa = 7.08TSRR4 pKa = 11.84RR5 pKa = 11.84SVKK8 pKa = 9.91SGPRR12 pKa = 11.84EE13 pKa = 3.94VPRR16 pKa = 11.84DD17 pKa = 3.7EE18 pKa = 5.67YY19 pKa = 11.42EE20 pKa = 4.01DD21 pKa = 4.8LYY23 pKa = 9.79YY24 pKa = 10.73TPSSGMASPDD34 pKa = 3.65SPPDD38 pKa = 3.3TSRR41 pKa = 11.84RR42 pKa = 11.84GALQTRR48 pKa = 11.84SRR50 pKa = 11.84QRR52 pKa = 11.84GEE54 pKa = 3.87VRR56 pKa = 11.84FVQYY60 pKa = 10.87DD61 pKa = 3.49EE62 pKa = 5.07SDD64 pKa = 3.58YY65 pKa = 11.75ALYY68 pKa = 10.61GGSSSEE74 pKa = 4.18DD75 pKa = 3.52DD76 pKa = 3.62EE77 pKa = 5.61HH78 pKa = 8.45PEE80 pKa = 3.91VPRR83 pKa = 11.84TRR85 pKa = 11.84RR86 pKa = 11.84PVSGAVLSGPGPARR100 pKa = 11.84APPPPAGSGGAGRR113 pKa = 11.84TPTTAPRR120 pKa = 11.84APRR123 pKa = 11.84TQRR126 pKa = 11.84VATKK130 pKa = 10.67APAAPAAEE138 pKa = 4.35TTRR141 pKa = 11.84GRR143 pKa = 11.84KK144 pKa = 8.6SAQPEE149 pKa = 4.16SAALPDD155 pKa = 4.48APASTAPTRR164 pKa = 11.84SKK166 pKa = 10.07TPAQGLARR174 pKa = 11.84KK175 pKa = 8.77LHH177 pKa = 6.51FSTAPPNPDD186 pKa = 3.71APWTPRR192 pKa = 11.84VAGFNKK198 pKa = 9.92RR199 pKa = 11.84VFCAAVGRR207 pKa = 11.84LAAMHH212 pKa = 6.39ARR214 pKa = 11.84MAAVQLWDD222 pKa = 3.36MSRR225 pKa = 11.84PRR227 pKa = 11.84TDD229 pKa = 3.27EE230 pKa = 4.46DD231 pKa = 3.98LNEE234 pKa = 4.24LLGITTIRR242 pKa = 11.84VTVCEE247 pKa = 4.57GKK249 pKa = 10.09NLLQRR254 pKa = 11.84ANEE257 pKa = 4.1LVNPDD262 pKa = 3.04VVQDD266 pKa = 3.4VDD268 pKa = 3.24AATATRR274 pKa = 11.84GRR276 pKa = 11.84SAASRR281 pKa = 11.84PTEE284 pKa = 4.06RR285 pKa = 11.84PRR287 pKa = 11.84APARR291 pKa = 11.84SASRR295 pKa = 11.84PRR297 pKa = 11.84RR298 pKa = 11.84PVEE301 pKa = 3.64
MM1 pKa = 7.08TSRR4 pKa = 11.84RR5 pKa = 11.84SVKK8 pKa = 9.91SGPRR12 pKa = 11.84EE13 pKa = 3.94VPRR16 pKa = 11.84DD17 pKa = 3.7EE18 pKa = 5.67YY19 pKa = 11.42EE20 pKa = 4.01DD21 pKa = 4.8LYY23 pKa = 9.79YY24 pKa = 10.73TPSSGMASPDD34 pKa = 3.65SPPDD38 pKa = 3.3TSRR41 pKa = 11.84RR42 pKa = 11.84GALQTRR48 pKa = 11.84SRR50 pKa = 11.84QRR52 pKa = 11.84GEE54 pKa = 3.87VRR56 pKa = 11.84FVQYY60 pKa = 10.87DD61 pKa = 3.49EE62 pKa = 5.07SDD64 pKa = 3.58YY65 pKa = 11.75ALYY68 pKa = 10.61GGSSSEE74 pKa = 4.18DD75 pKa = 3.52DD76 pKa = 3.62EE77 pKa = 5.61HH78 pKa = 8.45PEE80 pKa = 3.91VPRR83 pKa = 11.84TRR85 pKa = 11.84RR86 pKa = 11.84PVSGAVLSGPGPARR100 pKa = 11.84APPPPAGSGGAGRR113 pKa = 11.84TPTTAPRR120 pKa = 11.84APRR123 pKa = 11.84TQRR126 pKa = 11.84VATKK130 pKa = 10.67APAAPAAEE138 pKa = 4.35TTRR141 pKa = 11.84GRR143 pKa = 11.84KK144 pKa = 8.6SAQPEE149 pKa = 4.16SAALPDD155 pKa = 4.48APASTAPTRR164 pKa = 11.84SKK166 pKa = 10.07TPAQGLARR174 pKa = 11.84KK175 pKa = 8.77LHH177 pKa = 6.51FSTAPPNPDD186 pKa = 3.71APWTPRR192 pKa = 11.84VAGFNKK198 pKa = 9.92RR199 pKa = 11.84VFCAAVGRR207 pKa = 11.84LAAMHH212 pKa = 6.39ARR214 pKa = 11.84MAAVQLWDD222 pKa = 3.36MSRR225 pKa = 11.84PRR227 pKa = 11.84TDD229 pKa = 3.27EE230 pKa = 4.46DD231 pKa = 3.98LNEE234 pKa = 4.24LLGITTIRR242 pKa = 11.84VTVCEE247 pKa = 4.57GKK249 pKa = 10.09NLLQRR254 pKa = 11.84ANEE257 pKa = 4.1LVNPDD262 pKa = 3.04VVQDD266 pKa = 3.4VDD268 pKa = 3.24AATATRR274 pKa = 11.84GRR276 pKa = 11.84SAASRR281 pKa = 11.84PTEE284 pKa = 4.06RR285 pKa = 11.84PRR287 pKa = 11.84APARR291 pKa = 11.84SASRR295 pKa = 11.84PRR297 pKa = 11.84RR298 pKa = 11.84PVEE301 pKa = 3.64
Molecular weight: 32.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
39997 |
88 |
3164 |
519.4 |
56.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.538 ± 0.449 | 1.815 ± 0.112 |
5.308 ± 0.13 | 4.838 ± 0.135 |
3.41 ± 0.162 | 7.873 ± 0.201 |
2.528 ± 0.099 | 2.85 ± 0.157 |
1.758 ± 0.128 | 9.683 ± 0.26 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.72 ± 0.102 | 2.293 ± 0.148 |
8.918 ± 0.363 | 3.083 ± 0.142 |
8.406 ± 0.201 | 6.238 ± 0.209 |
6.013 ± 0.198 | 7.026 ± 0.185 |
1.158 ± 0.069 | 2.545 ± 0.138 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
