
Ferrithrix thermotolerans DSM 19514
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Acidimicrobiia; Acidimicrobiales; Acidimicrobiaceae; Ferrithrix; Ferrithrix thermotolerans
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
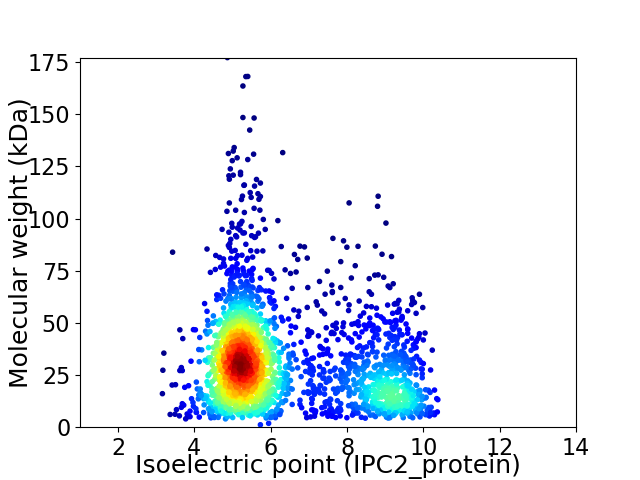
Virtual 2D-PAGE plot for 2339 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4WJM3|A0A1M4WJM3_9ACTN DNA polymerase-3 subunit epsilon OS=Ferrithrix thermotolerans DSM 19514 OX=1121881 GN=SAMN02745225_01695 PE=4 SV=1
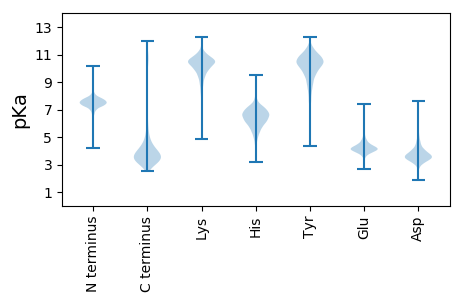
MM1 pKa = 7.07TRR3 pKa = 11.84SLSAAISGIDD13 pKa = 4.66ASQAWLDD20 pKa = 3.83NIGNNIANVDD30 pKa = 3.74TVGYY34 pKa = 9.89QSTDD38 pKa = 2.67IQFADD43 pKa = 4.72LLYY46 pKa = 10.38QQQQSAGAPQVGVNGGTNPIEE67 pKa = 4.34IGSGVRR73 pKa = 11.84VASTGINFTQGTLSQTGVQTDD94 pKa = 3.95LAIQGQGFLVVQQGGNTFYY113 pKa = 10.88TRR115 pKa = 11.84AGNLQLDD122 pKa = 3.99AAGEE126 pKa = 4.22LVTPSGAIVQGWTPNAAGVINTSGPLTNLTIPQGQAANPTATKK169 pKa = 10.49NINLGGNLPAWNGSGTAPSYY189 pKa = 9.33TATVTAFDD197 pKa = 4.06SLGGQIPITLTFTASTTPNTWSVVATAPNPAGGTDD232 pKa = 3.28TLTPAGTTISFNGTTGQMTSTAPITLSGFTGYY264 pKa = 11.31NNLPAGYY271 pKa = 8.7TMNLDD276 pKa = 4.19FPAAGTPQAVTQFSGTSTIQVTSQDD301 pKa = 3.64GNASGTLDD309 pKa = 3.45GFTISSNGEE318 pKa = 3.53IVGNYY323 pKa = 10.18SNGNTQPLGQIALAQFANDD342 pKa = 3.07QGLAKK347 pKa = 10.38VGNLLYY353 pKa = 10.5QATLNSGAPQLGLAGSGGLGTLVGGAVEE381 pKa = 4.2TSNVNLGTQLTDD393 pKa = 4.77LIVAQTAYY401 pKa = 10.23QANTKK406 pKa = 9.63VVSTTSTVLQSLVQMAA422 pKa = 4.2
MM1 pKa = 7.07TRR3 pKa = 11.84SLSAAISGIDD13 pKa = 4.66ASQAWLDD20 pKa = 3.83NIGNNIANVDD30 pKa = 3.74TVGYY34 pKa = 9.89QSTDD38 pKa = 2.67IQFADD43 pKa = 4.72LLYY46 pKa = 10.38QQQQSAGAPQVGVNGGTNPIEE67 pKa = 4.34IGSGVRR73 pKa = 11.84VASTGINFTQGTLSQTGVQTDD94 pKa = 3.95LAIQGQGFLVVQQGGNTFYY113 pKa = 10.88TRR115 pKa = 11.84AGNLQLDD122 pKa = 3.99AAGEE126 pKa = 4.22LVTPSGAIVQGWTPNAAGVINTSGPLTNLTIPQGQAANPTATKK169 pKa = 10.49NINLGGNLPAWNGSGTAPSYY189 pKa = 9.33TATVTAFDD197 pKa = 4.06SLGGQIPITLTFTASTTPNTWSVVATAPNPAGGTDD232 pKa = 3.28TLTPAGTTISFNGTTGQMTSTAPITLSGFTGYY264 pKa = 11.31NNLPAGYY271 pKa = 8.7TMNLDD276 pKa = 4.19FPAAGTPQAVTQFSGTSTIQVTSQDD301 pKa = 3.64GNASGTLDD309 pKa = 3.45GFTISSNGEE318 pKa = 3.53IVGNYY323 pKa = 10.18SNGNTQPLGQIALAQFANDD342 pKa = 3.07QGLAKK347 pKa = 10.38VGNLLYY353 pKa = 10.5QATLNSGAPQLGLAGSGGLGTLVGGAVEE381 pKa = 4.2TSNVNLGTQLTDD393 pKa = 4.77LIVAQTAYY401 pKa = 10.23QANTKK406 pKa = 9.63VVSTTSTVLQSLVQMAA422 pKa = 4.2
Molecular weight: 42.45 kDa
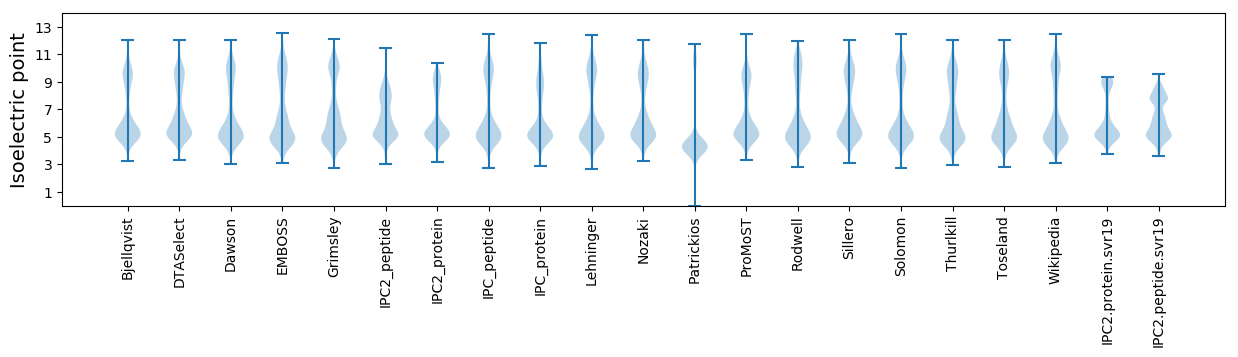
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4XH81|A0A1M4XH81_9ACTN Fur family transcriptional regulator ferric uptake regulator OS=Ferrithrix thermotolerans DSM 19514 OX=1121881 GN=SAMN02745225_02021 PE=3 SV=1
MM1 pKa = 7.26NVHH4 pKa = 6.65LVDD7 pKa = 3.6QAAFDD12 pKa = 4.4AVDD15 pKa = 3.15TCVRR19 pKa = 11.84HH20 pKa = 5.73LASIVATSSLKK31 pKa = 9.95AKK33 pKa = 9.58IWRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 7.69PNGAEE43 pKa = 3.29RR44 pKa = 11.84HH45 pKa = 4.49YY46 pKa = 11.16AYY48 pKa = 10.58ACLEE52 pKa = 4.01RR53 pKa = 11.84SSALGIIMRR62 pKa = 11.84GGTPDD67 pKa = 2.4ITMRR71 pKa = 11.84QHH73 pKa = 6.87SLTKK77 pKa = 10.12KK78 pKa = 7.99EE79 pKa = 3.96RR80 pKa = 11.84KK81 pKa = 9.61AIASYY86 pKa = 9.15LHH88 pKa = 6.19RR89 pKa = 11.84ALRR92 pKa = 11.84KK93 pKa = 9.61ALAHH97 pKa = 5.81TWPTVRR103 pKa = 11.84LVRR106 pKa = 11.84SMALDD111 pKa = 3.36EE112 pKa = 4.3TLYY115 pKa = 10.94SSFVIEE121 pKa = 4.48RR122 pKa = 11.84PGKK125 pKa = 7.78EE126 pKa = 4.22HH127 pKa = 7.36PGKK130 pKa = 8.66TPKK133 pKa = 9.85RR134 pKa = 11.84RR135 pKa = 11.84QYY137 pKa = 10.2VQVIGPRR144 pKa = 11.84PRR146 pKa = 11.84EE147 pKa = 4.19RR148 pKa = 11.84ITIPLSGVSRR158 pKa = 11.84VSGNIRR164 pKa = 11.84VVLDD168 pKa = 3.73EE169 pKa = 4.38GSTRR173 pKa = 11.84AFIHH177 pKa = 5.43VAYY180 pKa = 9.52EE181 pKa = 3.96IQPLPGPATGPDD193 pKa = 3.3VAIDD197 pKa = 3.24WGITEE202 pKa = 4.2VCTDD206 pKa = 3.77DD207 pKa = 5.46SGIKK211 pKa = 10.13HH212 pKa = 5.76GASYY216 pKa = 10.9GAILKK221 pKa = 10.27RR222 pKa = 11.84ATEE225 pKa = 3.85QRR227 pKa = 11.84KK228 pKa = 7.88RR229 pKa = 11.84TGQVRR234 pKa = 11.84NKK236 pKa = 9.66LWAITKK242 pKa = 9.88KK243 pKa = 10.67DD244 pKa = 3.59AGSKK248 pKa = 8.17RR249 pKa = 11.84AKK251 pKa = 10.33RR252 pKa = 11.84IAKK255 pKa = 9.94YY256 pKa = 10.79NLGTKK261 pKa = 9.06KK262 pKa = 9.51QARR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84ARR269 pKa = 11.84TQAALQTVAGAAIKK283 pKa = 9.97EE284 pKa = 4.28VIYY287 pKa = 11.18GEE289 pKa = 4.33GNRR292 pKa = 11.84TRR294 pKa = 11.84ARR296 pKa = 11.84GKK298 pKa = 9.88VPQLPAHH305 pKa = 6.8RR306 pKa = 11.84PRR308 pKa = 11.84RR309 pKa = 11.84VIVEE313 pKa = 4.41DD314 pKa = 3.88LAQLRR319 pKa = 11.84GKK321 pKa = 10.2AKK323 pKa = 9.83SKK325 pKa = 10.49KK326 pKa = 9.46ISRR329 pKa = 11.84LCSSWARR336 pKa = 11.84SEE338 pKa = 4.05NEE340 pKa = 3.1EE341 pKa = 3.97RR342 pKa = 11.84MAVHH346 pKa = 7.17AYY348 pKa = 9.43IGRR351 pKa = 11.84SDD353 pKa = 3.61VRR355 pKa = 11.84VVSAAYY361 pKa = 8.85TSQTCPDD368 pKa = 3.31PHH370 pKa = 7.87YY371 pKa = 10.96GYY373 pKa = 10.95VSQDD377 pKa = 2.96NRR379 pKa = 11.84SGDD382 pKa = 3.26WFHH385 pKa = 6.98CRR387 pKa = 11.84NPYY390 pKa = 8.79WEE392 pKa = 5.02CNRR395 pKa = 11.84QGDD398 pKa = 4.32ADD400 pKa = 3.99HH401 pKa = 6.49VAAVNLKK408 pKa = 8.76TRR410 pKa = 11.84INDD413 pKa = 3.72PEE415 pKa = 3.87IHH417 pKa = 6.97RR418 pKa = 11.84FTPHH422 pKa = 7.43IEE424 pKa = 3.94VKK426 pKa = 10.49KK427 pKa = 10.54ILDD430 pKa = 3.27ARR432 pKa = 11.84FVCRR436 pKa = 11.84RR437 pKa = 11.84EE438 pKa = 3.88SRR440 pKa = 11.84TKK442 pKa = 10.97GGDD445 pKa = 3.04ASPASRR451 pKa = 11.84GQRR454 pKa = 11.84DD455 pKa = 3.45ATSTSGAQGASVDD468 pKa = 4.12GAATAHH474 pKa = 6.03GRR476 pKa = 11.84TSIPRR481 pKa = 11.84RR482 pKa = 11.84SDD484 pKa = 3.19PDD486 pKa = 3.43VGGGTTTDD494 pKa = 3.03LHH496 pKa = 6.2RR497 pKa = 11.84VRR499 pKa = 11.84SIWRR503 pKa = 11.84GPGRR507 pKa = 11.84LSSARR512 pKa = 11.84AKK514 pKa = 9.58TKK516 pKa = 10.2GVPSVPLQKK525 pKa = 9.86FTEE528 pKa = 4.31KK529 pKa = 10.38QQ530 pKa = 3.06
MM1 pKa = 7.26NVHH4 pKa = 6.65LVDD7 pKa = 3.6QAAFDD12 pKa = 4.4AVDD15 pKa = 3.15TCVRR19 pKa = 11.84HH20 pKa = 5.73LASIVATSSLKK31 pKa = 9.95AKK33 pKa = 9.58IWRR36 pKa = 11.84RR37 pKa = 11.84YY38 pKa = 7.69PNGAEE43 pKa = 3.29RR44 pKa = 11.84HH45 pKa = 4.49YY46 pKa = 11.16AYY48 pKa = 10.58ACLEE52 pKa = 4.01RR53 pKa = 11.84SSALGIIMRR62 pKa = 11.84GGTPDD67 pKa = 2.4ITMRR71 pKa = 11.84QHH73 pKa = 6.87SLTKK77 pKa = 10.12KK78 pKa = 7.99EE79 pKa = 3.96RR80 pKa = 11.84KK81 pKa = 9.61AIASYY86 pKa = 9.15LHH88 pKa = 6.19RR89 pKa = 11.84ALRR92 pKa = 11.84KK93 pKa = 9.61ALAHH97 pKa = 5.81TWPTVRR103 pKa = 11.84LVRR106 pKa = 11.84SMALDD111 pKa = 3.36EE112 pKa = 4.3TLYY115 pKa = 10.94SSFVIEE121 pKa = 4.48RR122 pKa = 11.84PGKK125 pKa = 7.78EE126 pKa = 4.22HH127 pKa = 7.36PGKK130 pKa = 8.66TPKK133 pKa = 9.85RR134 pKa = 11.84RR135 pKa = 11.84QYY137 pKa = 10.2VQVIGPRR144 pKa = 11.84PRR146 pKa = 11.84EE147 pKa = 4.19RR148 pKa = 11.84ITIPLSGVSRR158 pKa = 11.84VSGNIRR164 pKa = 11.84VVLDD168 pKa = 3.73EE169 pKa = 4.38GSTRR173 pKa = 11.84AFIHH177 pKa = 5.43VAYY180 pKa = 9.52EE181 pKa = 3.96IQPLPGPATGPDD193 pKa = 3.3VAIDD197 pKa = 3.24WGITEE202 pKa = 4.2VCTDD206 pKa = 3.77DD207 pKa = 5.46SGIKK211 pKa = 10.13HH212 pKa = 5.76GASYY216 pKa = 10.9GAILKK221 pKa = 10.27RR222 pKa = 11.84ATEE225 pKa = 3.85QRR227 pKa = 11.84KK228 pKa = 7.88RR229 pKa = 11.84TGQVRR234 pKa = 11.84NKK236 pKa = 9.66LWAITKK242 pKa = 9.88KK243 pKa = 10.67DD244 pKa = 3.59AGSKK248 pKa = 8.17RR249 pKa = 11.84AKK251 pKa = 10.33RR252 pKa = 11.84IAKK255 pKa = 9.94YY256 pKa = 10.79NLGTKK261 pKa = 9.06KK262 pKa = 9.51QARR265 pKa = 11.84RR266 pKa = 11.84RR267 pKa = 11.84ARR269 pKa = 11.84TQAALQTVAGAAIKK283 pKa = 9.97EE284 pKa = 4.28VIYY287 pKa = 11.18GEE289 pKa = 4.33GNRR292 pKa = 11.84TRR294 pKa = 11.84ARR296 pKa = 11.84GKK298 pKa = 9.88VPQLPAHH305 pKa = 6.8RR306 pKa = 11.84PRR308 pKa = 11.84RR309 pKa = 11.84VIVEE313 pKa = 4.41DD314 pKa = 3.88LAQLRR319 pKa = 11.84GKK321 pKa = 10.2AKK323 pKa = 9.83SKK325 pKa = 10.49KK326 pKa = 9.46ISRR329 pKa = 11.84LCSSWARR336 pKa = 11.84SEE338 pKa = 4.05NEE340 pKa = 3.1EE341 pKa = 3.97RR342 pKa = 11.84MAVHH346 pKa = 7.17AYY348 pKa = 9.43IGRR351 pKa = 11.84SDD353 pKa = 3.61VRR355 pKa = 11.84VVSAAYY361 pKa = 8.85TSQTCPDD368 pKa = 3.31PHH370 pKa = 7.87YY371 pKa = 10.96GYY373 pKa = 10.95VSQDD377 pKa = 2.96NRR379 pKa = 11.84SGDD382 pKa = 3.26WFHH385 pKa = 6.98CRR387 pKa = 11.84NPYY390 pKa = 8.79WEE392 pKa = 5.02CNRR395 pKa = 11.84QGDD398 pKa = 4.32ADD400 pKa = 3.99HH401 pKa = 6.49VAAVNLKK408 pKa = 8.76TRR410 pKa = 11.84INDD413 pKa = 3.72PEE415 pKa = 3.87IHH417 pKa = 6.97RR418 pKa = 11.84FTPHH422 pKa = 7.43IEE424 pKa = 3.94VKK426 pKa = 10.49KK427 pKa = 10.54ILDD430 pKa = 3.27ARR432 pKa = 11.84FVCRR436 pKa = 11.84RR437 pKa = 11.84EE438 pKa = 3.88SRR440 pKa = 11.84TKK442 pKa = 10.97GGDD445 pKa = 3.04ASPASRR451 pKa = 11.84GQRR454 pKa = 11.84DD455 pKa = 3.45ATSTSGAQGASVDD468 pKa = 4.12GAATAHH474 pKa = 6.03GRR476 pKa = 11.84TSIPRR481 pKa = 11.84RR482 pKa = 11.84SDD484 pKa = 3.19PDD486 pKa = 3.43VGGGTTTDD494 pKa = 3.03LHH496 pKa = 6.2RR497 pKa = 11.84VRR499 pKa = 11.84SIWRR503 pKa = 11.84GPGRR507 pKa = 11.84LSSARR512 pKa = 11.84AKK514 pKa = 9.58TKK516 pKa = 10.2GVPSVPLQKK525 pKa = 9.86FTEE528 pKa = 4.31KK529 pKa = 10.38QQ530 pKa = 3.06
Molecular weight: 58.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
706337 |
10 |
1597 |
302.0 |
33.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.016 ± 0.049 | 0.843 ± 0.017 |
5.313 ± 0.045 | 6.482 ± 0.056 |
3.742 ± 0.029 | 7.808 ± 0.05 |
1.929 ± 0.021 | 5.304 ± 0.034 |
4.177 ± 0.031 | 10.572 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.017 | 2.802 ± 0.03 |
4.54 ± 0.036 | 3.092 ± 0.038 |
6.142 ± 0.051 | 8.03 ± 0.057 |
5.395 ± 0.043 | 8.737 ± 0.051 |
1.11 ± 0.021 | 2.863 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
