
Trypanosoma cruzi (strain CL Brener)
Taxonomy: cellular organisms; Eukaryota; Discoba; Euglenozoa; Kinetoplastea; Metakinetoplastina; Trypanosomatida; Trypanosomatidae; Trypanosoma; Schizotrypanum; Trypanosoma cruzi
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
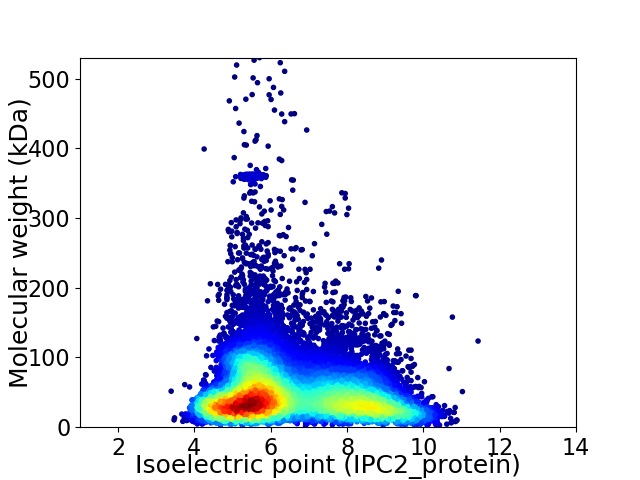
Virtual 2D-PAGE plot for 19243 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q4DH69|Q4DH69_TRYCC Uncharacterized protein OS=Trypanosoma cruzi (strain CL Brener) OX=353153 GN=Tc00.1047053509527.140 PE=4 SV=1
MM1 pKa = 8.07DD2 pKa = 5.65GYY4 pKa = 11.61SLLEE8 pKa = 3.98RR9 pKa = 11.84LDD11 pKa = 4.05AFFSEE16 pKa = 5.14GEE18 pKa = 4.06NTDD21 pKa = 3.77AIGNFLSEE29 pKa = 4.44EE30 pKa = 4.01QGVMQLLGQPTDD42 pKa = 3.42SQEE45 pKa = 4.03ALEE48 pKa = 4.99FYY50 pKa = 11.15SLFKK54 pKa = 10.55RR55 pKa = 11.84YY56 pKa = 9.75AVVVDD61 pKa = 3.6NLLNAFIEE69 pKa = 4.49RR70 pKa = 11.84EE71 pKa = 4.21SKK73 pKa = 10.38LGYY76 pKa = 10.72VIDD79 pKa = 4.64LEE81 pKa = 4.22QLAAAVMNEE90 pKa = 3.34WHH92 pKa = 6.29QEE94 pKa = 3.62QDD96 pKa = 2.83FCRR99 pKa = 11.84YY100 pKa = 9.26VCTAYY105 pKa = 9.86IAGALDD111 pKa = 3.59FDD113 pKa = 4.48SFKK116 pKa = 11.22QLVADD121 pKa = 4.12VNAITAYY128 pKa = 9.92PFGDD132 pKa = 3.72EE133 pKa = 4.9SSDD136 pKa = 3.36ADD138 pKa = 4.37SVTEE142 pKa = 4.0TNTQEE147 pKa = 4.08EE148 pKa = 5.06EE149 pKa = 3.89II150 pKa = 4.62
MM1 pKa = 8.07DD2 pKa = 5.65GYY4 pKa = 11.61SLLEE8 pKa = 3.98RR9 pKa = 11.84LDD11 pKa = 4.05AFFSEE16 pKa = 5.14GEE18 pKa = 4.06NTDD21 pKa = 3.77AIGNFLSEE29 pKa = 4.44EE30 pKa = 4.01QGVMQLLGQPTDD42 pKa = 3.42SQEE45 pKa = 4.03ALEE48 pKa = 4.99FYY50 pKa = 11.15SLFKK54 pKa = 10.55RR55 pKa = 11.84YY56 pKa = 9.75AVVVDD61 pKa = 3.6NLLNAFIEE69 pKa = 4.49RR70 pKa = 11.84EE71 pKa = 4.21SKK73 pKa = 10.38LGYY76 pKa = 10.72VIDD79 pKa = 4.64LEE81 pKa = 4.22QLAAAVMNEE90 pKa = 3.34WHH92 pKa = 6.29QEE94 pKa = 3.62QDD96 pKa = 2.83FCRR99 pKa = 11.84YY100 pKa = 9.26VCTAYY105 pKa = 9.86IAGALDD111 pKa = 3.59FDD113 pKa = 4.48SFKK116 pKa = 11.22QLVADD121 pKa = 4.12VNAITAYY128 pKa = 9.92PFGDD132 pKa = 3.72EE133 pKa = 4.9SSDD136 pKa = 3.36ADD138 pKa = 4.37SVTEE142 pKa = 4.0TNTQEE147 pKa = 4.08EE148 pKa = 5.06EE149 pKa = 3.89II150 pKa = 4.62
Molecular weight: 16.87 kDa
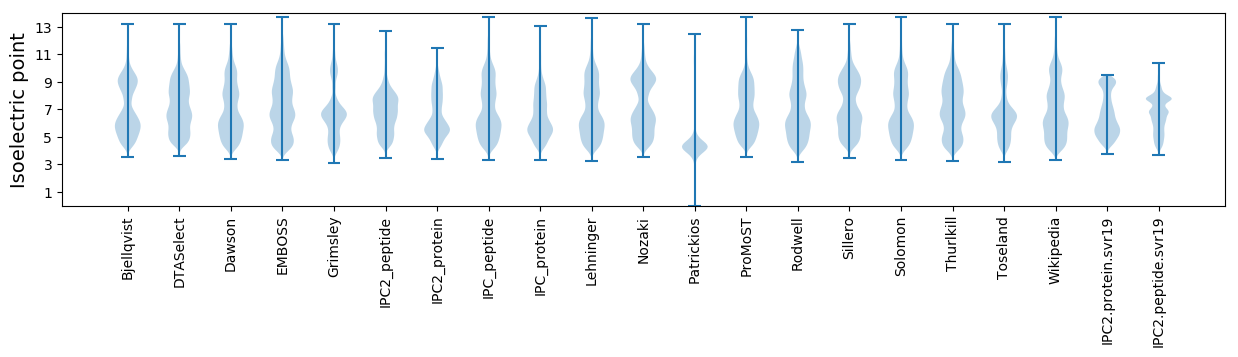
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q4D504|Q4D504_TRYCC Ras-related protein rab-5 putative (Fragment) OS=Trypanosoma cruzi (strain CL Brener) OX=353153 GN=Tc00.1047053511269.4 PE=4 SV=1
RR1 pKa = 7.48RR2 pKa = 11.84RR3 pKa = 11.84LRR5 pKa = 11.84SAWRR9 pKa = 11.84RR10 pKa = 11.84SWRR13 pKa = 11.84CARR16 pKa = 11.84TTSKK20 pKa = 10.71SVQTTSKK27 pKa = 10.85SVLLLPRR34 pKa = 11.84MPLAVAVPLQGRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84SAWRR55 pKa = 11.84RR56 pKa = 11.84SWRR59 pKa = 11.84NARR62 pKa = 11.84TTSKK66 pKa = 10.85SVLLLPRR73 pKa = 11.84MPLAVAVPLQGRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84LRR90 pKa = 11.84SAWRR94 pKa = 11.84RR95 pKa = 11.84SWRR98 pKa = 11.84CARR101 pKa = 11.84TTSKK105 pKa = 10.65SVQTISKK112 pKa = 10.23SVLLLPRR119 pKa = 11.84MPLAVAVPLQGRR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LRR136 pKa = 11.84SAWRR140 pKa = 11.84RR141 pKa = 11.84SWRR144 pKa = 11.84NARR147 pKa = 11.84TTSKK151 pKa = 10.85SVLLLPRR158 pKa = 11.84MPLAVAVPLQGRR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84LRR175 pKa = 11.84SAWRR179 pKa = 11.84RR180 pKa = 11.84SWRR183 pKa = 11.84NARR186 pKa = 11.84TTSKK190 pKa = 10.85SVLLLPRR197 pKa = 11.84MPLAVAVPLQGRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84LRR214 pKa = 11.84SAWRR218 pKa = 11.84RR219 pKa = 11.84SWRR222 pKa = 11.84NARR225 pKa = 11.84TTSKK229 pKa = 10.85SVLLLPRR236 pKa = 11.84MPLAVAVPLQGRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84LRR253 pKa = 11.84SAWRR257 pKa = 11.84RR258 pKa = 11.84SWRR261 pKa = 11.84CARR264 pKa = 11.84TTSKK268 pKa = 10.71SVQTTSKK275 pKa = 10.85SVLLLPRR282 pKa = 11.84MPLAVAVPLQGRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84LRR299 pKa = 11.84SAWRR303 pKa = 11.84RR304 pKa = 11.84SWRR307 pKa = 11.84NARR310 pKa = 11.84TTSKK314 pKa = 10.85SVLLLPRR321 pKa = 11.84MPLAVAVPLQGRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84LRR338 pKa = 11.84SAWRR342 pKa = 11.84RR343 pKa = 11.84SWRR346 pKa = 11.84CARR349 pKa = 11.84TTSKK353 pKa = 10.65SVQTISKK360 pKa = 10.23SVLLLPRR367 pKa = 11.84MPLAVAVPLQGRR379 pKa = 11.84RR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84LRR384 pKa = 11.84SAWRR388 pKa = 11.84RR389 pKa = 11.84SWRR392 pKa = 11.84NARR395 pKa = 11.84TTSKK399 pKa = 10.85SVLLLPRR406 pKa = 11.84MPLAVAVPLQGRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LRR423 pKa = 11.84SAWRR427 pKa = 11.84RR428 pKa = 11.84SWRR431 pKa = 11.84NARR434 pKa = 11.84TTSKK438 pKa = 10.85SVLLLPRR445 pKa = 11.84MPLAVAVPLQGRR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84LRR462 pKa = 11.84SAWRR466 pKa = 11.84RR467 pKa = 11.84SWRR470 pKa = 11.84CARR473 pKa = 11.84TTSKK477 pKa = 10.71SVQTTSKK484 pKa = 10.85SVLLLPRR491 pKa = 11.84MPLAVAVPLQGRR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84LRR508 pKa = 11.84SAWRR512 pKa = 11.84RR513 pKa = 11.84SWRR516 pKa = 11.84CARR519 pKa = 11.84TTSKK523 pKa = 10.71SVQTTSKK530 pKa = 10.85SVLLLPRR537 pKa = 11.84MPFAVAVPLQGRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84RR552 pKa = 11.84LRR554 pKa = 11.84SAWRR558 pKa = 11.84RR559 pKa = 11.84SWRR562 pKa = 11.84CARR565 pKa = 11.84TTSKK569 pKa = 10.71SVQTTSKK576 pKa = 10.85SVLLLPRR583 pKa = 11.84MPLAVAVPLQGRR595 pKa = 11.84RR596 pKa = 11.84RR597 pKa = 11.84RR598 pKa = 11.84LRR600 pKa = 11.84SAWKK604 pKa = 10.23RR605 pKa = 11.84SWRR608 pKa = 11.84CARR611 pKa = 11.84TTSKK615 pKa = 10.88SVLLLPRR622 pKa = 11.84MPLAVAVPLQGRR634 pKa = 11.84RR635 pKa = 11.84RR636 pKa = 11.84RR637 pKa = 11.84LRR639 pKa = 11.84SAWRR643 pKa = 11.84RR644 pKa = 11.84SWRR647 pKa = 11.84CARR650 pKa = 11.84TTSKK654 pKa = 10.88SVLLLPRR661 pKa = 11.84MPLAVAVPLQGRR673 pKa = 11.84RR674 pKa = 11.84RR675 pKa = 11.84RR676 pKa = 11.84LRR678 pKa = 11.84SAWRR682 pKa = 11.84RR683 pKa = 11.84SWRR686 pKa = 11.84CARR689 pKa = 11.84TTSKK693 pKa = 10.88SVLLLPRR700 pKa = 11.84MPLAVAVPLQGRR712 pKa = 11.84RR713 pKa = 11.84RR714 pKa = 11.84RR715 pKa = 11.84LRR717 pKa = 11.84SAWRR721 pKa = 11.84RR722 pKa = 11.84SWRR725 pKa = 11.84CARR728 pKa = 11.84TTSKK732 pKa = 10.88SVLLLPRR739 pKa = 11.84MPLAVAVPLQGRR751 pKa = 11.84RR752 pKa = 11.84RR753 pKa = 11.84RR754 pKa = 11.84LRR756 pKa = 11.84SAWRR760 pKa = 11.84RR761 pKa = 11.84SWRR764 pKa = 11.84CARR767 pKa = 11.84TTSKK771 pKa = 10.88SVLLLPRR778 pKa = 11.84MPLAVAVPLQGRR790 pKa = 11.84RR791 pKa = 11.84RR792 pKa = 11.84RR793 pKa = 11.84LRR795 pKa = 11.84SAWRR799 pKa = 11.84RR800 pKa = 11.84SWRR803 pKa = 11.84NARR806 pKa = 11.84TTSKK810 pKa = 10.85SVLLLPRR817 pKa = 11.84MPLAVAVPLQGRR829 pKa = 11.84RR830 pKa = 11.84RR831 pKa = 11.84RR832 pKa = 11.84LRR834 pKa = 11.84SAWRR838 pKa = 11.84RR839 pKa = 11.84SWRR842 pKa = 11.84CARR845 pKa = 11.84TISKK849 pKa = 10.31SVLLLPRR856 pKa = 11.84MPLAVAVPLQGRR868 pKa = 11.84RR869 pKa = 11.84RR870 pKa = 11.84RR871 pKa = 11.84LRR873 pKa = 11.84SAWRR877 pKa = 11.84RR878 pKa = 11.84SWRR881 pKa = 11.84NARR884 pKa = 11.84TTSKK888 pKa = 10.85SVLLLPRR895 pKa = 11.84MPLAVAVPLQGRR907 pKa = 11.84RR908 pKa = 11.84RR909 pKa = 11.84RR910 pKa = 11.84LRR912 pKa = 11.84SAWRR916 pKa = 11.84RR917 pKa = 11.84SWRR920 pKa = 11.84NARR923 pKa = 11.84TTSKK927 pKa = 10.85SVLLLPRR934 pKa = 11.84MPLAVAVPLQGRR946 pKa = 11.84RR947 pKa = 11.84RR948 pKa = 11.84RR949 pKa = 11.84LRR951 pKa = 11.84SAWRR955 pKa = 11.84RR956 pKa = 11.84SWRR959 pKa = 11.84NARR962 pKa = 11.84TTSKK966 pKa = 10.85SVLLLPRR973 pKa = 11.84MPLAVAVPLQGRR985 pKa = 11.84RR986 pKa = 11.84RR987 pKa = 11.84RR988 pKa = 11.84LRR990 pKa = 11.84SAWKK994 pKa = 10.23RR995 pKa = 11.84SWRR998 pKa = 11.84CARR1001 pKa = 11.84TTSKK1005 pKa = 10.71SVQTTSKK1012 pKa = 10.87SVLLPSMMPLQGRR1025 pKa = 11.84RR1026 pKa = 11.84RR1027 pKa = 11.84RR1028 pKa = 11.84LRR1030 pKa = 11.84SAWRR1034 pKa = 11.84RR1035 pKa = 11.84SWRR1038 pKa = 11.84NARR1041 pKa = 11.84TTSRR1045 pKa = 11.84IMWHH1049 pKa = 5.86RR1050 pKa = 11.84WW1051 pKa = 2.87
RR1 pKa = 7.48RR2 pKa = 11.84RR3 pKa = 11.84LRR5 pKa = 11.84SAWRR9 pKa = 11.84RR10 pKa = 11.84SWRR13 pKa = 11.84CARR16 pKa = 11.84TTSKK20 pKa = 10.71SVQTTSKK27 pKa = 10.85SVLLLPRR34 pKa = 11.84MPLAVAVPLQGRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84LRR51 pKa = 11.84SAWRR55 pKa = 11.84RR56 pKa = 11.84SWRR59 pKa = 11.84NARR62 pKa = 11.84TTSKK66 pKa = 10.85SVLLLPRR73 pKa = 11.84MPLAVAVPLQGRR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84LRR90 pKa = 11.84SAWRR94 pKa = 11.84RR95 pKa = 11.84SWRR98 pKa = 11.84CARR101 pKa = 11.84TTSKK105 pKa = 10.65SVQTISKK112 pKa = 10.23SVLLLPRR119 pKa = 11.84MPLAVAVPLQGRR131 pKa = 11.84RR132 pKa = 11.84RR133 pKa = 11.84RR134 pKa = 11.84LRR136 pKa = 11.84SAWRR140 pKa = 11.84RR141 pKa = 11.84SWRR144 pKa = 11.84NARR147 pKa = 11.84TTSKK151 pKa = 10.85SVLLLPRR158 pKa = 11.84MPLAVAVPLQGRR170 pKa = 11.84RR171 pKa = 11.84RR172 pKa = 11.84RR173 pKa = 11.84LRR175 pKa = 11.84SAWRR179 pKa = 11.84RR180 pKa = 11.84SWRR183 pKa = 11.84NARR186 pKa = 11.84TTSKK190 pKa = 10.85SVLLLPRR197 pKa = 11.84MPLAVAVPLQGRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84LRR214 pKa = 11.84SAWRR218 pKa = 11.84RR219 pKa = 11.84SWRR222 pKa = 11.84NARR225 pKa = 11.84TTSKK229 pKa = 10.85SVLLLPRR236 pKa = 11.84MPLAVAVPLQGRR248 pKa = 11.84RR249 pKa = 11.84RR250 pKa = 11.84RR251 pKa = 11.84LRR253 pKa = 11.84SAWRR257 pKa = 11.84RR258 pKa = 11.84SWRR261 pKa = 11.84CARR264 pKa = 11.84TTSKK268 pKa = 10.71SVQTTSKK275 pKa = 10.85SVLLLPRR282 pKa = 11.84MPLAVAVPLQGRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84RR297 pKa = 11.84LRR299 pKa = 11.84SAWRR303 pKa = 11.84RR304 pKa = 11.84SWRR307 pKa = 11.84NARR310 pKa = 11.84TTSKK314 pKa = 10.85SVLLLPRR321 pKa = 11.84MPLAVAVPLQGRR333 pKa = 11.84RR334 pKa = 11.84RR335 pKa = 11.84RR336 pKa = 11.84LRR338 pKa = 11.84SAWRR342 pKa = 11.84RR343 pKa = 11.84SWRR346 pKa = 11.84CARR349 pKa = 11.84TTSKK353 pKa = 10.65SVQTISKK360 pKa = 10.23SVLLLPRR367 pKa = 11.84MPLAVAVPLQGRR379 pKa = 11.84RR380 pKa = 11.84RR381 pKa = 11.84RR382 pKa = 11.84LRR384 pKa = 11.84SAWRR388 pKa = 11.84RR389 pKa = 11.84SWRR392 pKa = 11.84NARR395 pKa = 11.84TTSKK399 pKa = 10.85SVLLLPRR406 pKa = 11.84MPLAVAVPLQGRR418 pKa = 11.84RR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84LRR423 pKa = 11.84SAWRR427 pKa = 11.84RR428 pKa = 11.84SWRR431 pKa = 11.84NARR434 pKa = 11.84TTSKK438 pKa = 10.85SVLLLPRR445 pKa = 11.84MPLAVAVPLQGRR457 pKa = 11.84RR458 pKa = 11.84RR459 pKa = 11.84RR460 pKa = 11.84LRR462 pKa = 11.84SAWRR466 pKa = 11.84RR467 pKa = 11.84SWRR470 pKa = 11.84CARR473 pKa = 11.84TTSKK477 pKa = 10.71SVQTTSKK484 pKa = 10.85SVLLLPRR491 pKa = 11.84MPLAVAVPLQGRR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84RR506 pKa = 11.84LRR508 pKa = 11.84SAWRR512 pKa = 11.84RR513 pKa = 11.84SWRR516 pKa = 11.84CARR519 pKa = 11.84TTSKK523 pKa = 10.71SVQTTSKK530 pKa = 10.85SVLLLPRR537 pKa = 11.84MPFAVAVPLQGRR549 pKa = 11.84RR550 pKa = 11.84RR551 pKa = 11.84RR552 pKa = 11.84LRR554 pKa = 11.84SAWRR558 pKa = 11.84RR559 pKa = 11.84SWRR562 pKa = 11.84CARR565 pKa = 11.84TTSKK569 pKa = 10.71SVQTTSKK576 pKa = 10.85SVLLLPRR583 pKa = 11.84MPLAVAVPLQGRR595 pKa = 11.84RR596 pKa = 11.84RR597 pKa = 11.84RR598 pKa = 11.84LRR600 pKa = 11.84SAWKK604 pKa = 10.23RR605 pKa = 11.84SWRR608 pKa = 11.84CARR611 pKa = 11.84TTSKK615 pKa = 10.88SVLLLPRR622 pKa = 11.84MPLAVAVPLQGRR634 pKa = 11.84RR635 pKa = 11.84RR636 pKa = 11.84RR637 pKa = 11.84LRR639 pKa = 11.84SAWRR643 pKa = 11.84RR644 pKa = 11.84SWRR647 pKa = 11.84CARR650 pKa = 11.84TTSKK654 pKa = 10.88SVLLLPRR661 pKa = 11.84MPLAVAVPLQGRR673 pKa = 11.84RR674 pKa = 11.84RR675 pKa = 11.84RR676 pKa = 11.84LRR678 pKa = 11.84SAWRR682 pKa = 11.84RR683 pKa = 11.84SWRR686 pKa = 11.84CARR689 pKa = 11.84TTSKK693 pKa = 10.88SVLLLPRR700 pKa = 11.84MPLAVAVPLQGRR712 pKa = 11.84RR713 pKa = 11.84RR714 pKa = 11.84RR715 pKa = 11.84LRR717 pKa = 11.84SAWRR721 pKa = 11.84RR722 pKa = 11.84SWRR725 pKa = 11.84CARR728 pKa = 11.84TTSKK732 pKa = 10.88SVLLLPRR739 pKa = 11.84MPLAVAVPLQGRR751 pKa = 11.84RR752 pKa = 11.84RR753 pKa = 11.84RR754 pKa = 11.84LRR756 pKa = 11.84SAWRR760 pKa = 11.84RR761 pKa = 11.84SWRR764 pKa = 11.84CARR767 pKa = 11.84TTSKK771 pKa = 10.88SVLLLPRR778 pKa = 11.84MPLAVAVPLQGRR790 pKa = 11.84RR791 pKa = 11.84RR792 pKa = 11.84RR793 pKa = 11.84LRR795 pKa = 11.84SAWRR799 pKa = 11.84RR800 pKa = 11.84SWRR803 pKa = 11.84NARR806 pKa = 11.84TTSKK810 pKa = 10.85SVLLLPRR817 pKa = 11.84MPLAVAVPLQGRR829 pKa = 11.84RR830 pKa = 11.84RR831 pKa = 11.84RR832 pKa = 11.84LRR834 pKa = 11.84SAWRR838 pKa = 11.84RR839 pKa = 11.84SWRR842 pKa = 11.84CARR845 pKa = 11.84TISKK849 pKa = 10.31SVLLLPRR856 pKa = 11.84MPLAVAVPLQGRR868 pKa = 11.84RR869 pKa = 11.84RR870 pKa = 11.84RR871 pKa = 11.84LRR873 pKa = 11.84SAWRR877 pKa = 11.84RR878 pKa = 11.84SWRR881 pKa = 11.84NARR884 pKa = 11.84TTSKK888 pKa = 10.85SVLLLPRR895 pKa = 11.84MPLAVAVPLQGRR907 pKa = 11.84RR908 pKa = 11.84RR909 pKa = 11.84RR910 pKa = 11.84LRR912 pKa = 11.84SAWRR916 pKa = 11.84RR917 pKa = 11.84SWRR920 pKa = 11.84NARR923 pKa = 11.84TTSKK927 pKa = 10.85SVLLLPRR934 pKa = 11.84MPLAVAVPLQGRR946 pKa = 11.84RR947 pKa = 11.84RR948 pKa = 11.84RR949 pKa = 11.84LRR951 pKa = 11.84SAWRR955 pKa = 11.84RR956 pKa = 11.84SWRR959 pKa = 11.84NARR962 pKa = 11.84TTSKK966 pKa = 10.85SVLLLPRR973 pKa = 11.84MPLAVAVPLQGRR985 pKa = 11.84RR986 pKa = 11.84RR987 pKa = 11.84RR988 pKa = 11.84LRR990 pKa = 11.84SAWKK994 pKa = 10.23RR995 pKa = 11.84SWRR998 pKa = 11.84CARR1001 pKa = 11.84TTSKK1005 pKa = 10.71SVQTTSKK1012 pKa = 10.87SVLLPSMMPLQGRR1025 pKa = 11.84RR1026 pKa = 11.84RR1027 pKa = 11.84RR1028 pKa = 11.84LRR1030 pKa = 11.84SAWRR1034 pKa = 11.84RR1035 pKa = 11.84SWRR1038 pKa = 11.84NARR1041 pKa = 11.84TTSRR1045 pKa = 11.84IMWHH1049 pKa = 5.86RR1050 pKa = 11.84WW1051 pKa = 2.87
Molecular weight: 123.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9671180 |
18 |
4848 |
502.6 |
55.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.812 ± 0.023 | 2.121 ± 0.01 |
4.858 ± 0.01 | 6.958 ± 0.028 |
3.63 ± 0.013 | 7.002 ± 0.025 |
2.444 ± 0.008 | 3.651 ± 0.014 |
4.263 ± 0.021 | 9.533 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.434 ± 0.007 | 3.552 ± 0.009 |
4.948 ± 0.015 | 3.627 ± 0.014 |
6.805 ± 0.019 | 8.12 ± 0.021 |
5.981 ± 0.016 | 7.718 ± 0.029 |
1.178 ± 0.006 | 2.364 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
