
Pseudomonas sp. HAR-UPW-AIA-41
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; unclassified Pseudomonas
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
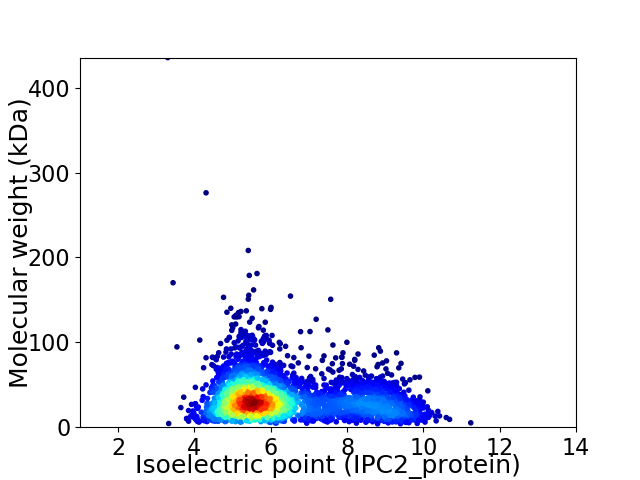
Virtual 2D-PAGE plot for 3419 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A2IR97|A0A2A2IR97_9PSED Ribosomal RNA large subunit methyltransferase G OS=Pseudomonas sp. HAR-UPW-AIA-41 OX=1985301 GN=rlmG PE=3 SV=1
GG1 pKa = 6.48QSAVLTIPYY10 pKa = 8.65TVTDD14 pKa = 3.92DD15 pKa = 3.31QGAISTANLVITVTGTNDD33 pKa = 3.05LSVISSASVDD43 pKa = 3.9VIEE46 pKa = 5.79SNASLNTGGTLSISDD61 pKa = 3.7IDD63 pKa = 4.11GPAPSFNPATNASGSGGYY81 pKa = 10.78GLFNLGSNGVWSYY94 pKa = 10.75STTGPLNQLVAGQSYY109 pKa = 9.9TDD111 pKa = 3.68TLTVTAADD119 pKa = 4.13GSTGTVTVTILGTNDD134 pKa = 3.03APVLGGVRR142 pKa = 11.84TSFTYY147 pKa = 10.37SEE149 pKa = 4.39SAASVITRR157 pKa = 11.84TLLDD161 pKa = 3.44SSVTLTDD168 pKa = 3.22IDD170 pKa = 3.92SSNFDD175 pKa = 3.41GGSVIVSISNGVATQDD191 pKa = 3.29QLTVQDD197 pKa = 3.76QGTGSGQISVVGSDD211 pKa = 2.91VRR213 pKa = 11.84YY214 pKa = 9.97NFGSGAVSIGTISGGTGGTPLTISLNSNATPVAVDD249 pKa = 3.61ALVQRR254 pKa = 11.84IAYY257 pKa = 8.21GNSSQLPTTTPRR269 pKa = 11.84TISITVNDD277 pKa = 4.12GDD279 pKa = 4.18GNANGGSSSVTQNITMNVTSVNDD302 pKa = 3.46APTFSNLGGARR313 pKa = 11.84AFTEE317 pKa = 4.17GGNAVVLDD325 pKa = 4.22NNASLGDD332 pKa = 3.65RR333 pKa = 11.84EE334 pKa = 4.34LAVLGDD340 pKa = 3.83FGGASLTLSRR350 pKa = 11.84RR351 pKa = 11.84GGTNSDD357 pKa = 3.28DD358 pKa = 4.09DD359 pKa = 4.22FRR361 pKa = 11.84GTGNLSLSAGEE372 pKa = 4.07VRR374 pKa = 11.84LSGVLVGSYY383 pKa = 10.52DD384 pKa = 3.8QAALTNGTLQITFNSGTSNAQANSVLRR411 pKa = 11.84QIAYY415 pKa = 10.74SNGSDD420 pKa = 3.6APPASVVIDD429 pKa = 3.5YY430 pKa = 11.05ALNDD434 pKa = 4.03NNSGAQGSGGAKK446 pKa = 8.64TATGSVTVNISAVNDD461 pKa = 3.61APVAGSQSNSTAEE474 pKa = 3.83DD475 pKa = 3.82APFNGQIIASDD486 pKa = 3.49VDD488 pKa = 4.09GGSLTYY494 pKa = 10.66SLQTGASNGSVTLNSSTGAFVYY516 pKa = 9.72TPNAGYY522 pKa = 10.4SGADD526 pKa = 3.01SFTVQVSDD534 pKa = 3.6GRR536 pKa = 11.84GGFVNSVINMSVTPVDD552 pKa = 3.92DD553 pKa = 4.85APVLSLTTPATPVEE567 pKa = 4.4GNAAEE572 pKa = 4.57GNLISTASATDD583 pKa = 3.32EE584 pKa = 4.17DD585 pKa = 5.11TPASGLNFSLTSNPGNVYY603 pKa = 10.46AINAATGAVTLTAAGAALVNAGGDD627 pKa = 3.75LPPVEE632 pKa = 4.42VTVTDD637 pKa = 4.25GNLTDD642 pKa = 3.83SGSMNVPATQDD653 pKa = 3.15INDD656 pKa = 4.24EE657 pKa = 4.09PTAIPLSLTTVEE669 pKa = 5.36DD670 pKa = 3.46NAIDD674 pKa = 4.08GKK676 pKa = 11.07VVASDD681 pKa = 4.05PDD683 pKa = 4.33GDD685 pKa = 3.95TLTYY689 pKa = 10.64SITTGNGPQHH699 pKa = 5.79GTVTLNPNGTFSYY712 pKa = 10.82LPTKK716 pKa = 10.37DD717 pKa = 3.83YY718 pKa = 11.62NGTDD722 pKa = 3.32AFTVTIDD729 pKa = 3.97DD730 pKa = 4.22GNGGVTISLVNITILPFNDD749 pKa = 3.47APEE752 pKa = 4.17TANQSKK758 pKa = 6.84TTNEE762 pKa = 4.04DD763 pKa = 3.05QSVSGQVIASDD774 pKa = 3.52VDD776 pKa = 3.57GDD778 pKa = 3.82RR779 pKa = 11.84LSYY782 pKa = 10.38IIQSGVANGSLLLNTATGEE801 pKa = 4.07YY802 pKa = 9.58TYY804 pKa = 10.82TPNKK808 pKa = 9.86DD809 pKa = 3.6YY810 pKa = 11.54NGTDD814 pKa = 3.12SFTLRR819 pKa = 11.84VYY821 pKa = 10.4DD822 pKa = 3.74PKK824 pKa = 11.11GGYY827 pKa = 7.4TEE829 pKa = 4.3SVVTITVAPVNDD841 pKa = 4.4DD842 pKa = 3.94PKK844 pKa = 10.96SSPLFVTTVEE854 pKa = 4.74DD855 pKa = 3.43NTATGKK861 pKa = 10.42INARR865 pKa = 11.84DD866 pKa = 3.51VDD868 pKa = 4.36GDD870 pKa = 3.81TLTYY874 pKa = 10.64SIATGNGPQHH884 pKa = 5.81GTVTLNPDD892 pKa = 3.2GTFSYY897 pKa = 10.84LPAKK901 pKa = 10.18DD902 pKa = 4.14YY903 pKa = 11.64NGTDD907 pKa = 3.22AFTVTIDD914 pKa = 3.97DD915 pKa = 4.26GNGGVTTSFVNITIQPFNDD934 pKa = 3.45APEE937 pKa = 4.16TANQSKK943 pKa = 6.84TTNEE947 pKa = 4.04DD948 pKa = 3.06QSVSGQIIASDD959 pKa = 3.51VDD961 pKa = 3.77GDD963 pKa = 3.76RR964 pKa = 11.84LNYY967 pKa = 9.84VIQSGVAHH975 pKa = 6.83GSILLNTVTGEE986 pKa = 3.92YY987 pKa = 9.84TYY989 pKa = 10.93TPNKK993 pKa = 9.91DD994 pKa = 3.55YY995 pKa = 11.56NGTDD999 pKa = 2.89TFTIRR1004 pKa = 11.84VYY1006 pKa = 10.63DD1007 pKa = 3.88PKK1009 pKa = 11.01GGYY1012 pKa = 7.53TDD1014 pKa = 3.85SVVTVTVAPVNDD1026 pKa = 4.33APTSGPLFMNTVEE1039 pKa = 5.54DD1040 pKa = 3.49IAANGTIITRR1050 pKa = 11.84DD1051 pKa = 3.13VDD1053 pKa = 4.03GGTLTYY1059 pKa = 10.67SIATGNGPQHH1069 pKa = 5.81GTVTLNPDD1077 pKa = 3.2GTFSYY1082 pKa = 10.84LPAKK1086 pKa = 10.18DD1087 pKa = 4.14YY1088 pKa = 11.64NGTDD1092 pKa = 3.22AFTVTIDD1099 pKa = 3.97DD1100 pKa = 4.26GNGGVTTSLVNITIQPFNDD1119 pKa = 3.45APEE1122 pKa = 4.16TANQSKK1128 pKa = 6.84TTNEE1132 pKa = 4.04DD1133 pKa = 3.06QSVSGQIIASDD1144 pKa = 3.5VDD1146 pKa = 3.95GDD1148 pKa = 4.17SLSYY1152 pKa = 10.57AIQSGATHH1160 pKa = 6.82GSILLYY1166 pKa = 9.86TATGEE1171 pKa = 3.93YY1172 pKa = 9.82TYY1174 pKa = 10.91RR1175 pKa = 11.84PNKK1178 pKa = 10.01DD1179 pKa = 3.37YY1180 pKa = 11.62NGTDD1184 pKa = 2.89TFTIRR1189 pKa = 11.84VYY1191 pKa = 10.63DD1192 pKa = 3.88PKK1194 pKa = 11.01GGYY1197 pKa = 7.53TDD1199 pKa = 3.85SVVTVTVNPVDD1210 pKa = 3.98DD1211 pKa = 5.08APILSLTTPTTPVEE1225 pKa = 4.22GSAAEE1230 pKa = 4.25GNLISTASATDD1241 pKa = 3.29KK1242 pKa = 9.68DD1243 pKa = 4.3TPATGLSFSLTNNPGNVYY1261 pKa = 10.38AINAATGAVTLTAAGAALVNAGGDD1285 pKa = 3.75LPPVEE1290 pKa = 4.47VTVTDD1295 pKa = 3.62GTTPISGSVNVPATSDD1311 pKa = 3.34INDD1314 pKa = 3.45APIINVTAQTLIEE1327 pKa = 4.11NSAAAGSVAATYY1339 pKa = 7.68TASDD1343 pKa = 3.91EE1344 pKa = 4.31EE1345 pKa = 4.26ASSFIVSFTPGSNDD1359 pKa = 3.04DD1360 pKa = 3.81GYY1362 pKa = 11.89YY1363 pKa = 10.32VLANGRR1369 pKa = 11.84VEE1371 pKa = 4.1LTQDD1375 pKa = 3.41GADD1378 pKa = 3.55HH1379 pKa = 6.51VNAGNSLPPVSLVVTEE1395 pKa = 4.41TGSPALTGTDD1405 pKa = 3.02SDD1407 pKa = 4.43TPNVTLTNDD1416 pKa = 3.32APVANADD1423 pKa = 3.65TASTNEE1429 pKa = 4.09EE1430 pKa = 4.24TPVTIDD1436 pKa = 3.38VLSNDD1441 pKa = 3.83SDD1443 pKa = 3.93VDD1445 pKa = 4.02GPTLSIKK1452 pKa = 10.34SASVDD1457 pKa = 3.53PAQGTVSIVAGQLVFTPASNFTGAATITYY1486 pKa = 9.59VSTDD1490 pKa = 3.23GSDD1493 pKa = 3.7DD1494 pKa = 3.72SAPATVTVTVNPVNDD1509 pKa = 3.63APVANTDD1516 pKa = 3.57TASTNEE1522 pKa = 4.19DD1523 pKa = 3.71TPVTIDD1529 pKa = 3.49VLSNDD1534 pKa = 3.7SDD1536 pKa = 3.85VDD1538 pKa = 4.24GPALSIKK1545 pKa = 10.11SASVDD1550 pKa = 3.53PAQGTVSIVAGQLVFTPASNFTGTATITYY1579 pKa = 9.37VSTDD1583 pKa = 3.23GSDD1586 pKa = 3.7DD1587 pKa = 3.72SAPATVTVTVNPINDD1602 pKa = 3.59APVAQASTASTGEE1615 pKa = 3.71NTAFNGNVPAASDD1628 pKa = 3.0IDD1630 pKa = 3.96GTVNPNGYY1638 pKa = 10.5ALVNGPAAGSLTFNANGSYY1657 pKa = 10.66SFNPDD1662 pKa = 2.77SAFDD1666 pKa = 3.99DD1667 pKa = 4.19LAAGEE1672 pKa = 4.41SRR1674 pKa = 11.84NVTFTYY1680 pKa = 9.74TATDD1684 pKa = 3.4NNN1686 pKa = 4.08
GG1 pKa = 6.48QSAVLTIPYY10 pKa = 8.65TVTDD14 pKa = 3.92DD15 pKa = 3.31QGAISTANLVITVTGTNDD33 pKa = 3.05LSVISSASVDD43 pKa = 3.9VIEE46 pKa = 5.79SNASLNTGGTLSISDD61 pKa = 3.7IDD63 pKa = 4.11GPAPSFNPATNASGSGGYY81 pKa = 10.78GLFNLGSNGVWSYY94 pKa = 10.75STTGPLNQLVAGQSYY109 pKa = 9.9TDD111 pKa = 3.68TLTVTAADD119 pKa = 4.13GSTGTVTVTILGTNDD134 pKa = 3.03APVLGGVRR142 pKa = 11.84TSFTYY147 pKa = 10.37SEE149 pKa = 4.39SAASVITRR157 pKa = 11.84TLLDD161 pKa = 3.44SSVTLTDD168 pKa = 3.22IDD170 pKa = 3.92SSNFDD175 pKa = 3.41GGSVIVSISNGVATQDD191 pKa = 3.29QLTVQDD197 pKa = 3.76QGTGSGQISVVGSDD211 pKa = 2.91VRR213 pKa = 11.84YY214 pKa = 9.97NFGSGAVSIGTISGGTGGTPLTISLNSNATPVAVDD249 pKa = 3.61ALVQRR254 pKa = 11.84IAYY257 pKa = 8.21GNSSQLPTTTPRR269 pKa = 11.84TISITVNDD277 pKa = 4.12GDD279 pKa = 4.18GNANGGSSSVTQNITMNVTSVNDD302 pKa = 3.46APTFSNLGGARR313 pKa = 11.84AFTEE317 pKa = 4.17GGNAVVLDD325 pKa = 4.22NNASLGDD332 pKa = 3.65RR333 pKa = 11.84EE334 pKa = 4.34LAVLGDD340 pKa = 3.83FGGASLTLSRR350 pKa = 11.84RR351 pKa = 11.84GGTNSDD357 pKa = 3.28DD358 pKa = 4.09DD359 pKa = 4.22FRR361 pKa = 11.84GTGNLSLSAGEE372 pKa = 4.07VRR374 pKa = 11.84LSGVLVGSYY383 pKa = 10.52DD384 pKa = 3.8QAALTNGTLQITFNSGTSNAQANSVLRR411 pKa = 11.84QIAYY415 pKa = 10.74SNGSDD420 pKa = 3.6APPASVVIDD429 pKa = 3.5YY430 pKa = 11.05ALNDD434 pKa = 4.03NNSGAQGSGGAKK446 pKa = 8.64TATGSVTVNISAVNDD461 pKa = 3.61APVAGSQSNSTAEE474 pKa = 3.83DD475 pKa = 3.82APFNGQIIASDD486 pKa = 3.49VDD488 pKa = 4.09GGSLTYY494 pKa = 10.66SLQTGASNGSVTLNSSTGAFVYY516 pKa = 9.72TPNAGYY522 pKa = 10.4SGADD526 pKa = 3.01SFTVQVSDD534 pKa = 3.6GRR536 pKa = 11.84GGFVNSVINMSVTPVDD552 pKa = 3.92DD553 pKa = 4.85APVLSLTTPATPVEE567 pKa = 4.4GNAAEE572 pKa = 4.57GNLISTASATDD583 pKa = 3.32EE584 pKa = 4.17DD585 pKa = 5.11TPASGLNFSLTSNPGNVYY603 pKa = 10.46AINAATGAVTLTAAGAALVNAGGDD627 pKa = 3.75LPPVEE632 pKa = 4.42VTVTDD637 pKa = 4.25GNLTDD642 pKa = 3.83SGSMNVPATQDD653 pKa = 3.15INDD656 pKa = 4.24EE657 pKa = 4.09PTAIPLSLTTVEE669 pKa = 5.36DD670 pKa = 3.46NAIDD674 pKa = 4.08GKK676 pKa = 11.07VVASDD681 pKa = 4.05PDD683 pKa = 4.33GDD685 pKa = 3.95TLTYY689 pKa = 10.64SITTGNGPQHH699 pKa = 5.79GTVTLNPNGTFSYY712 pKa = 10.82LPTKK716 pKa = 10.37DD717 pKa = 3.83YY718 pKa = 11.62NGTDD722 pKa = 3.32AFTVTIDD729 pKa = 3.97DD730 pKa = 4.22GNGGVTISLVNITILPFNDD749 pKa = 3.47APEE752 pKa = 4.17TANQSKK758 pKa = 6.84TTNEE762 pKa = 4.04DD763 pKa = 3.05QSVSGQVIASDD774 pKa = 3.52VDD776 pKa = 3.57GDD778 pKa = 3.82RR779 pKa = 11.84LSYY782 pKa = 10.38IIQSGVANGSLLLNTATGEE801 pKa = 4.07YY802 pKa = 9.58TYY804 pKa = 10.82TPNKK808 pKa = 9.86DD809 pKa = 3.6YY810 pKa = 11.54NGTDD814 pKa = 3.12SFTLRR819 pKa = 11.84VYY821 pKa = 10.4DD822 pKa = 3.74PKK824 pKa = 11.11GGYY827 pKa = 7.4TEE829 pKa = 4.3SVVTITVAPVNDD841 pKa = 4.4DD842 pKa = 3.94PKK844 pKa = 10.96SSPLFVTTVEE854 pKa = 4.74DD855 pKa = 3.43NTATGKK861 pKa = 10.42INARR865 pKa = 11.84DD866 pKa = 3.51VDD868 pKa = 4.36GDD870 pKa = 3.81TLTYY874 pKa = 10.64SIATGNGPQHH884 pKa = 5.81GTVTLNPDD892 pKa = 3.2GTFSYY897 pKa = 10.84LPAKK901 pKa = 10.18DD902 pKa = 4.14YY903 pKa = 11.64NGTDD907 pKa = 3.22AFTVTIDD914 pKa = 3.97DD915 pKa = 4.26GNGGVTTSFVNITIQPFNDD934 pKa = 3.45APEE937 pKa = 4.16TANQSKK943 pKa = 6.84TTNEE947 pKa = 4.04DD948 pKa = 3.06QSVSGQIIASDD959 pKa = 3.51VDD961 pKa = 3.77GDD963 pKa = 3.76RR964 pKa = 11.84LNYY967 pKa = 9.84VIQSGVAHH975 pKa = 6.83GSILLNTVTGEE986 pKa = 3.92YY987 pKa = 9.84TYY989 pKa = 10.93TPNKK993 pKa = 9.91DD994 pKa = 3.55YY995 pKa = 11.56NGTDD999 pKa = 2.89TFTIRR1004 pKa = 11.84VYY1006 pKa = 10.63DD1007 pKa = 3.88PKK1009 pKa = 11.01GGYY1012 pKa = 7.53TDD1014 pKa = 3.85SVVTVTVAPVNDD1026 pKa = 4.33APTSGPLFMNTVEE1039 pKa = 5.54DD1040 pKa = 3.49IAANGTIITRR1050 pKa = 11.84DD1051 pKa = 3.13VDD1053 pKa = 4.03GGTLTYY1059 pKa = 10.67SIATGNGPQHH1069 pKa = 5.81GTVTLNPDD1077 pKa = 3.2GTFSYY1082 pKa = 10.84LPAKK1086 pKa = 10.18DD1087 pKa = 4.14YY1088 pKa = 11.64NGTDD1092 pKa = 3.22AFTVTIDD1099 pKa = 3.97DD1100 pKa = 4.26GNGGVTTSLVNITIQPFNDD1119 pKa = 3.45APEE1122 pKa = 4.16TANQSKK1128 pKa = 6.84TTNEE1132 pKa = 4.04DD1133 pKa = 3.06QSVSGQIIASDD1144 pKa = 3.5VDD1146 pKa = 3.95GDD1148 pKa = 4.17SLSYY1152 pKa = 10.57AIQSGATHH1160 pKa = 6.82GSILLYY1166 pKa = 9.86TATGEE1171 pKa = 3.93YY1172 pKa = 9.82TYY1174 pKa = 10.91RR1175 pKa = 11.84PNKK1178 pKa = 10.01DD1179 pKa = 3.37YY1180 pKa = 11.62NGTDD1184 pKa = 2.89TFTIRR1189 pKa = 11.84VYY1191 pKa = 10.63DD1192 pKa = 3.88PKK1194 pKa = 11.01GGYY1197 pKa = 7.53TDD1199 pKa = 3.85SVVTVTVNPVDD1210 pKa = 3.98DD1211 pKa = 5.08APILSLTTPTTPVEE1225 pKa = 4.22GSAAEE1230 pKa = 4.25GNLISTASATDD1241 pKa = 3.29KK1242 pKa = 9.68DD1243 pKa = 4.3TPATGLSFSLTNNPGNVYY1261 pKa = 10.38AINAATGAVTLTAAGAALVNAGGDD1285 pKa = 3.75LPPVEE1290 pKa = 4.47VTVTDD1295 pKa = 3.62GTTPISGSVNVPATSDD1311 pKa = 3.34INDD1314 pKa = 3.45APIINVTAQTLIEE1327 pKa = 4.11NSAAAGSVAATYY1339 pKa = 7.68TASDD1343 pKa = 3.91EE1344 pKa = 4.31EE1345 pKa = 4.26ASSFIVSFTPGSNDD1359 pKa = 3.04DD1360 pKa = 3.81GYY1362 pKa = 11.89YY1363 pKa = 10.32VLANGRR1369 pKa = 11.84VEE1371 pKa = 4.1LTQDD1375 pKa = 3.41GADD1378 pKa = 3.55HH1379 pKa = 6.51VNAGNSLPPVSLVVTEE1395 pKa = 4.41TGSPALTGTDD1405 pKa = 3.02SDD1407 pKa = 4.43TPNVTLTNDD1416 pKa = 3.32APVANADD1423 pKa = 3.65TASTNEE1429 pKa = 4.09EE1430 pKa = 4.24TPVTIDD1436 pKa = 3.38VLSNDD1441 pKa = 3.83SDD1443 pKa = 3.93VDD1445 pKa = 4.02GPTLSIKK1452 pKa = 10.34SASVDD1457 pKa = 3.53PAQGTVSIVAGQLVFTPASNFTGAATITYY1486 pKa = 9.59VSTDD1490 pKa = 3.23GSDD1493 pKa = 3.7DD1494 pKa = 3.72SAPATVTVTVNPVNDD1509 pKa = 3.63APVANTDD1516 pKa = 3.57TASTNEE1522 pKa = 4.19DD1523 pKa = 3.71TPVTIDD1529 pKa = 3.49VLSNDD1534 pKa = 3.7SDD1536 pKa = 3.85VDD1538 pKa = 4.24GPALSIKK1545 pKa = 10.11SASVDD1550 pKa = 3.53PAQGTVSIVAGQLVFTPASNFTGTATITYY1579 pKa = 9.37VSTDD1583 pKa = 3.23GSDD1586 pKa = 3.7DD1587 pKa = 3.72SAPATVTVTVNPINDD1602 pKa = 3.59APVAQASTASTGEE1615 pKa = 3.71NTAFNGNVPAASDD1628 pKa = 3.0IDD1630 pKa = 3.96GTVNPNGYY1638 pKa = 10.5ALVNGPAAGSLTFNANGSYY1657 pKa = 10.66SFNPDD1662 pKa = 2.77SAFDD1666 pKa = 3.99DD1667 pKa = 4.19LAAGEE1672 pKa = 4.41SRR1674 pKa = 11.84NVTFTYY1680 pKa = 9.74TATDD1684 pKa = 3.4NNN1686 pKa = 4.08
Molecular weight: 170.21 kDa
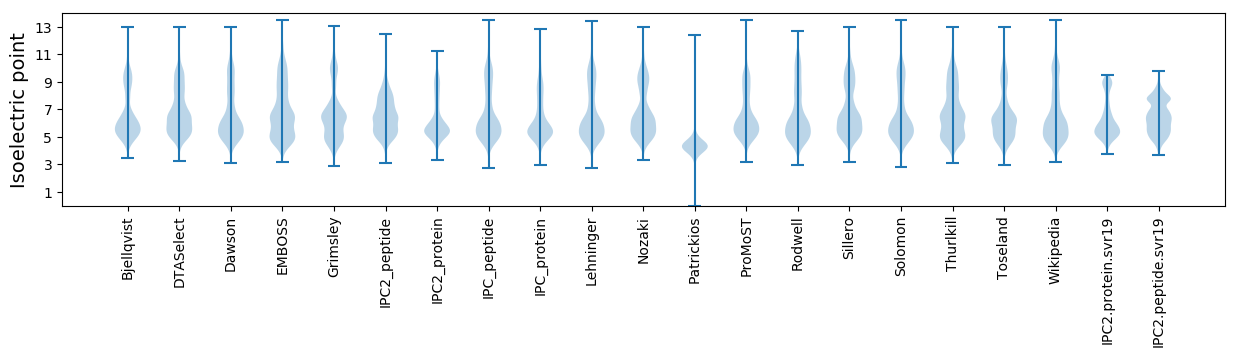
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A2ITS0|A0A2A2ITS0_9PSED Cell shape-determining protein MreC OS=Pseudomonas sp. HAR-UPW-AIA-41 OX=1985301 GN=CK486_11480 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1101474 |
33 |
4341 |
322.2 |
35.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.225 ± 0.052 | 1.042 ± 0.014 |
5.182 ± 0.029 | 6.097 ± 0.038 |
3.513 ± 0.025 | 7.843 ± 0.047 |
2.234 ± 0.021 | 4.52 ± 0.036 |
3.286 ± 0.038 | 12.536 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.199 ± 0.022 | 2.861 ± 0.028 |
4.842 ± 0.032 | 5.089 ± 0.039 |
6.652 ± 0.048 | 5.709 ± 0.034 |
4.4 ± 0.037 | 6.821 ± 0.038 |
1.45 ± 0.02 | 2.5 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
