
Pacific flying fox faeces associated circular DNA virus-12
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.56
Get precalculated fractions of proteins
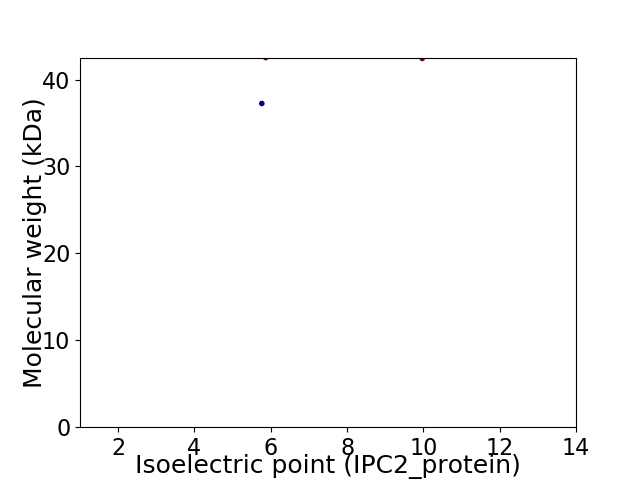
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
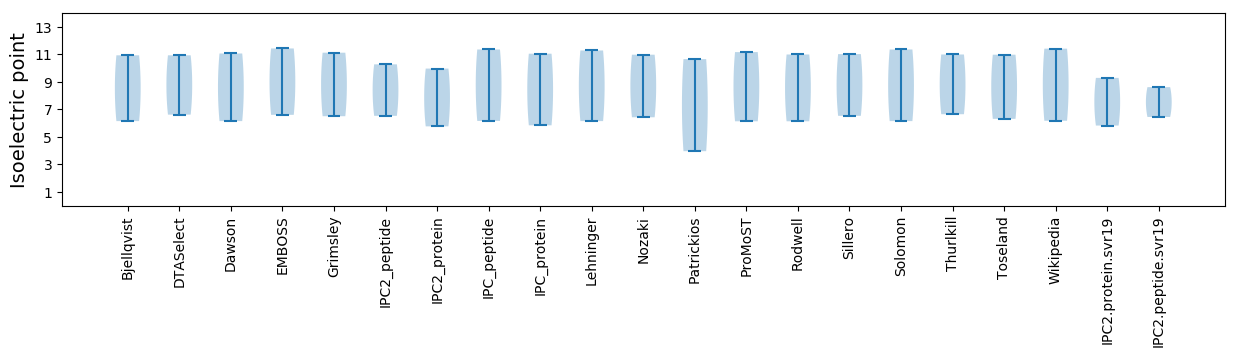
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTV5|A0A140CTV5_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-12 OX=1796007 PE=4 SV=1
MM1 pKa = 7.95QYY3 pKa = 9.94MDD5 pKa = 4.98DD6 pKa = 3.64AAADD10 pKa = 3.64VEE12 pKa = 4.45RR13 pKa = 11.84PDD15 pKa = 4.51IIAAQFRR22 pKa = 11.84LSAKK26 pKa = 10.18NLFLTYY32 pKa = 7.88PHH34 pKa = 7.0CGVQLEE40 pKa = 4.2IARR43 pKa = 11.84DD44 pKa = 3.73IILEE48 pKa = 3.9ALGRR52 pKa = 11.84GNVEE56 pKa = 4.08YY57 pKa = 10.12MVVAHH62 pKa = 6.55EE63 pKa = 4.08RR64 pKa = 11.84HH65 pKa = 5.6QDD67 pKa = 3.23GDD69 pKa = 3.87LHH71 pKa = 7.49IHH73 pKa = 6.39AVVCLIVRR81 pKa = 11.84CDD83 pKa = 3.13IRR85 pKa = 11.84NANLLDD91 pKa = 3.91LGEE94 pKa = 4.12YY95 pKa = 9.24HH96 pKa = 7.26GNYY99 pKa = 8.68QAIRR103 pKa = 11.84NVRR106 pKa = 11.84ASVRR110 pKa = 11.84YY111 pKa = 8.74VKK113 pKa = 10.38KK114 pKa = 10.51DD115 pKa = 3.32NVFIEE120 pKa = 4.34YY121 pKa = 8.75GTINAKK127 pKa = 10.27YY128 pKa = 10.64ADD130 pKa = 4.17AEE132 pKa = 4.1DD133 pKa = 3.92RR134 pKa = 11.84AAVLAKK140 pKa = 10.32LVQCTSEE147 pKa = 3.84ADD149 pKa = 3.26AMHH152 pKa = 7.05FIYY155 pKa = 11.11VNGHH159 pKa = 5.02ARR161 pKa = 11.84EE162 pKa = 4.33SYY164 pKa = 11.18LLLQYY169 pKa = 9.98WKK171 pKa = 8.97TKK173 pKa = 9.77QGEE176 pKa = 4.61SKK178 pKa = 10.38PGARR182 pKa = 11.84FARR185 pKa = 11.84DD186 pKa = 3.05AFLPCASLLAALEE199 pKa = 4.03EE200 pKa = 4.43LEE202 pKa = 4.44EE203 pKa = 4.5SDD205 pKa = 5.06RR206 pKa = 11.84CLLLIGPSGWGKK218 pKa = 6.87TQYY221 pKa = 11.48LLAHH225 pKa = 5.6YY226 pKa = 9.51NEE228 pKa = 4.26RR229 pKa = 11.84QIVRR233 pKa = 11.84ATHH236 pKa = 6.14MDD238 pKa = 3.75DD239 pKa = 3.61LKK241 pKa = 11.11KK242 pKa = 10.35ISKK245 pKa = 6.0TTEE248 pKa = 3.32ILILDD253 pKa = 4.3DD254 pKa = 4.54LSYY257 pKa = 11.5GHH259 pKa = 7.78LPRR262 pKa = 11.84QTLLHH267 pKa = 6.41LFDD270 pKa = 4.24MKK272 pKa = 10.37TDD274 pKa = 3.43RR275 pKa = 11.84SVHH278 pKa = 4.36VRR280 pKa = 11.84YY281 pKa = 7.65TVANLHH287 pKa = 5.82QSLLVILVGNEE298 pKa = 3.72LEE300 pKa = 4.5LVLGAHH306 pKa = 6.62IEE308 pKa = 4.29DD309 pKa = 3.82VAVMRR314 pKa = 11.84RR315 pKa = 11.84VTVFQLPHH323 pKa = 6.76RR324 pKa = 11.84LWDD327 pKa = 3.48
MM1 pKa = 7.95QYY3 pKa = 9.94MDD5 pKa = 4.98DD6 pKa = 3.64AAADD10 pKa = 3.64VEE12 pKa = 4.45RR13 pKa = 11.84PDD15 pKa = 4.51IIAAQFRR22 pKa = 11.84LSAKK26 pKa = 10.18NLFLTYY32 pKa = 7.88PHH34 pKa = 7.0CGVQLEE40 pKa = 4.2IARR43 pKa = 11.84DD44 pKa = 3.73IILEE48 pKa = 3.9ALGRR52 pKa = 11.84GNVEE56 pKa = 4.08YY57 pKa = 10.12MVVAHH62 pKa = 6.55EE63 pKa = 4.08RR64 pKa = 11.84HH65 pKa = 5.6QDD67 pKa = 3.23GDD69 pKa = 3.87LHH71 pKa = 7.49IHH73 pKa = 6.39AVVCLIVRR81 pKa = 11.84CDD83 pKa = 3.13IRR85 pKa = 11.84NANLLDD91 pKa = 3.91LGEE94 pKa = 4.12YY95 pKa = 9.24HH96 pKa = 7.26GNYY99 pKa = 8.68QAIRR103 pKa = 11.84NVRR106 pKa = 11.84ASVRR110 pKa = 11.84YY111 pKa = 8.74VKK113 pKa = 10.38KK114 pKa = 10.51DD115 pKa = 3.32NVFIEE120 pKa = 4.34YY121 pKa = 8.75GTINAKK127 pKa = 10.27YY128 pKa = 10.64ADD130 pKa = 4.17AEE132 pKa = 4.1DD133 pKa = 3.92RR134 pKa = 11.84AAVLAKK140 pKa = 10.32LVQCTSEE147 pKa = 3.84ADD149 pKa = 3.26AMHH152 pKa = 7.05FIYY155 pKa = 11.11VNGHH159 pKa = 5.02ARR161 pKa = 11.84EE162 pKa = 4.33SYY164 pKa = 11.18LLLQYY169 pKa = 9.98WKK171 pKa = 8.97TKK173 pKa = 9.77QGEE176 pKa = 4.61SKK178 pKa = 10.38PGARR182 pKa = 11.84FARR185 pKa = 11.84DD186 pKa = 3.05AFLPCASLLAALEE199 pKa = 4.03EE200 pKa = 4.43LEE202 pKa = 4.44EE203 pKa = 4.5SDD205 pKa = 5.06RR206 pKa = 11.84CLLLIGPSGWGKK218 pKa = 6.87TQYY221 pKa = 11.48LLAHH225 pKa = 5.6YY226 pKa = 9.51NEE228 pKa = 4.26RR229 pKa = 11.84QIVRR233 pKa = 11.84ATHH236 pKa = 6.14MDD238 pKa = 3.75DD239 pKa = 3.61LKK241 pKa = 11.11KK242 pKa = 10.35ISKK245 pKa = 6.0TTEE248 pKa = 3.32ILILDD253 pKa = 4.3DD254 pKa = 4.54LSYY257 pKa = 11.5GHH259 pKa = 7.78LPRR262 pKa = 11.84QTLLHH267 pKa = 6.41LFDD270 pKa = 4.24MKK272 pKa = 10.37TDD274 pKa = 3.43RR275 pKa = 11.84SVHH278 pKa = 4.36VRR280 pKa = 11.84YY281 pKa = 7.65TVANLHH287 pKa = 5.82QSLLVILVGNEE298 pKa = 3.72LEE300 pKa = 4.5LVLGAHH306 pKa = 6.62IEE308 pKa = 4.29DD309 pKa = 3.82VAVMRR314 pKa = 11.84RR315 pKa = 11.84VTVFQLPHH323 pKa = 6.76RR324 pKa = 11.84LWDD327 pKa = 3.48
Molecular weight: 37.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTV5|A0A140CTV5_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-12 OX=1796007 PE=4 SV=1
MM1 pKa = 7.95PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84ALTSSRR14 pKa = 11.84RR15 pKa = 11.84VRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TQRR22 pKa = 11.84RR23 pKa = 11.84GGRR26 pKa = 11.84GRR28 pKa = 11.84GLRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84SFRR36 pKa = 11.84RR37 pKa = 11.84GIRR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84GGFRR48 pKa = 11.84SRR50 pKa = 11.84RR51 pKa = 11.84NTGLSYY57 pKa = 10.92SVNITGWTSALLSWRR72 pKa = 11.84QSDD75 pKa = 3.76DD76 pKa = 2.9ACYY79 pKa = 10.72DD80 pKa = 3.6FLRR83 pKa = 11.84TEE85 pKa = 4.24HH86 pKa = 6.96RR87 pKa = 11.84PFINPEE93 pKa = 4.33LIGDD97 pKa = 4.07SNLEE101 pKa = 3.99VMGVANTTASQPAIPTKK118 pKa = 10.25SLVMGWRR125 pKa = 11.84DD126 pKa = 3.31QMSQFGRR133 pKa = 11.84NYY135 pKa = 8.86RR136 pKa = 11.84WGYY139 pKa = 9.1LKK141 pKa = 10.31SVRR144 pKa = 11.84YY145 pKa = 9.69RR146 pKa = 11.84IEE148 pKa = 3.3WCGTVTEE155 pKa = 4.61ATSYY159 pKa = 10.95MNGFMGWNASDD170 pKa = 3.87PGNVQGNQQGTIVTPRR186 pKa = 11.84WDD188 pKa = 3.55RR189 pKa = 11.84PFVNLLFSKK198 pKa = 10.06VHH200 pKa = 6.36EE201 pKa = 4.45EE202 pKa = 3.6FDD204 pKa = 3.39YY205 pKa = 11.3HH206 pKa = 7.5RR207 pKa = 11.84NQTGTAQLEE216 pKa = 4.3PFQNLGLGQPTRR228 pKa = 11.84LNYY231 pKa = 9.53PDD233 pKa = 3.7TMSSFQYY240 pKa = 8.03TTGGQVGSMMWSQLLNNRR258 pKa = 11.84AIKK261 pKa = 10.21KK262 pKa = 9.99LRR264 pKa = 11.84FTPRR268 pKa = 11.84DD269 pKa = 3.2NVKK272 pKa = 8.49WVKK275 pKa = 8.55WVPMTRR281 pKa = 11.84ADD283 pKa = 3.64KK284 pKa = 10.54MYY286 pKa = 11.09VKK288 pKa = 10.87YY289 pKa = 10.49DD290 pKa = 3.77LQKK293 pKa = 10.25MLQQTATPSPQYY305 pKa = 10.28YY306 pKa = 10.82DD307 pKa = 2.53MDD309 pKa = 3.93MRR311 pKa = 11.84VGGCWMAQEE320 pKa = 4.52RR321 pKa = 11.84PMAGAQVLGQTVDD334 pKa = 3.36VSISSNDD341 pKa = 3.21LFRR344 pKa = 11.84ITVYY348 pKa = 11.01SDD350 pKa = 2.35WKK352 pKa = 10.56FFGRR356 pKa = 11.84MQCFNTPLAA365 pKa = 4.57
MM1 pKa = 7.95PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84ALTSSRR14 pKa = 11.84RR15 pKa = 11.84VRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84TQRR22 pKa = 11.84RR23 pKa = 11.84GGRR26 pKa = 11.84GRR28 pKa = 11.84GLRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84SFRR36 pKa = 11.84RR37 pKa = 11.84GIRR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84RR44 pKa = 11.84GGFRR48 pKa = 11.84SRR50 pKa = 11.84RR51 pKa = 11.84NTGLSYY57 pKa = 10.92SVNITGWTSALLSWRR72 pKa = 11.84QSDD75 pKa = 3.76DD76 pKa = 2.9ACYY79 pKa = 10.72DD80 pKa = 3.6FLRR83 pKa = 11.84TEE85 pKa = 4.24HH86 pKa = 6.96RR87 pKa = 11.84PFINPEE93 pKa = 4.33LIGDD97 pKa = 4.07SNLEE101 pKa = 3.99VMGVANTTASQPAIPTKK118 pKa = 10.25SLVMGWRR125 pKa = 11.84DD126 pKa = 3.31QMSQFGRR133 pKa = 11.84NYY135 pKa = 8.86RR136 pKa = 11.84WGYY139 pKa = 9.1LKK141 pKa = 10.31SVRR144 pKa = 11.84YY145 pKa = 9.69RR146 pKa = 11.84IEE148 pKa = 3.3WCGTVTEE155 pKa = 4.61ATSYY159 pKa = 10.95MNGFMGWNASDD170 pKa = 3.87PGNVQGNQQGTIVTPRR186 pKa = 11.84WDD188 pKa = 3.55RR189 pKa = 11.84PFVNLLFSKK198 pKa = 10.06VHH200 pKa = 6.36EE201 pKa = 4.45EE202 pKa = 3.6FDD204 pKa = 3.39YY205 pKa = 11.3HH206 pKa = 7.5RR207 pKa = 11.84NQTGTAQLEE216 pKa = 4.3PFQNLGLGQPTRR228 pKa = 11.84LNYY231 pKa = 9.53PDD233 pKa = 3.7TMSSFQYY240 pKa = 8.03TTGGQVGSMMWSQLLNNRR258 pKa = 11.84AIKK261 pKa = 10.21KK262 pKa = 9.99LRR264 pKa = 11.84FTPRR268 pKa = 11.84DD269 pKa = 3.2NVKK272 pKa = 8.49WVKK275 pKa = 8.55WVPMTRR281 pKa = 11.84ADD283 pKa = 3.64KK284 pKa = 10.54MYY286 pKa = 11.09VKK288 pKa = 10.87YY289 pKa = 10.49DD290 pKa = 3.77LQKK293 pKa = 10.25MLQQTATPSPQYY305 pKa = 10.28YY306 pKa = 10.82DD307 pKa = 2.53MDD309 pKa = 3.93MRR311 pKa = 11.84VGGCWMAQEE320 pKa = 4.52RR321 pKa = 11.84PMAGAQVLGQTVDD334 pKa = 3.36VSISSNDD341 pKa = 3.21LFRR344 pKa = 11.84ITVYY348 pKa = 11.01SDD350 pKa = 2.35WKK352 pKa = 10.56FFGRR356 pKa = 11.84MQCFNTPLAA365 pKa = 4.57
Molecular weight: 42.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
692 |
327 |
365 |
346.0 |
39.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.081 ± 1.962 | 1.445 ± 0.254 |
5.78 ± 0.817 | 4.191 ± 1.255 |
3.468 ± 0.666 | 6.936 ± 1.332 |
2.746 ± 1.399 | 4.335 ± 0.961 |
3.468 ± 0.331 | 9.682 ± 2.26 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.468 ± 0.865 | 4.48 ± 0.528 |
3.468 ± 0.865 | 5.202 ± 0.799 |
9.971 ± 1.914 | 5.491 ± 1.187 |
5.636 ± 1.281 | 6.792 ± 0.755 |
2.168 ± 0.815 | 4.191 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
