
Candidatus Nitrosocosmicus franklandus
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrososphaeria; Nitrososphaerales; Nitrososphaeraceae; Candidatus Nitrosocosmicus
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
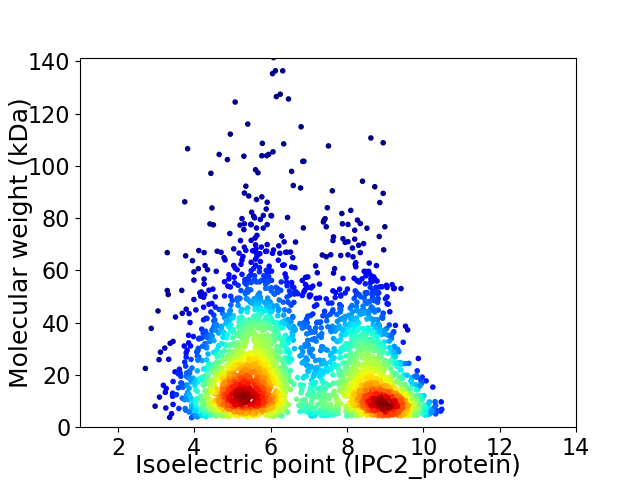
Virtual 2D-PAGE plot for 3135 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A484I8D6|A0A484I8D6_9ARCH Acetyl-coenzyme A synthetase OS=Candidatus Nitrosocosmicus franklandus OX=1798806 GN=acsA PE=3 SV=1
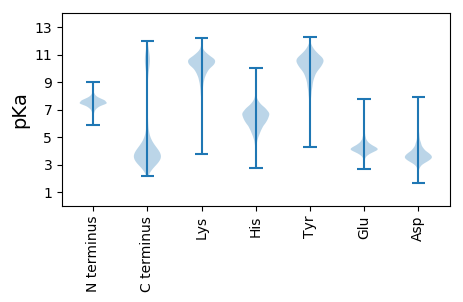
MM1 pKa = 7.4SLHH4 pKa = 6.33NEE6 pKa = 3.69NMIKK10 pKa = 10.24EE11 pKa = 4.29FSLLLILGILIVTSSTSMSNPIDD34 pKa = 3.81AQLNSSDD41 pKa = 3.91TNIGNNGVMSIGSSSGFSNTNTTGMGIDD69 pKa = 5.87DD70 pKa = 4.01IDD72 pKa = 3.52SWDD75 pKa = 3.66VVSGNWINRR84 pKa = 11.84AFTIQGGLPEE94 pKa = 4.22NSGLNRR100 pKa = 11.84IILYY104 pKa = 9.76PEE106 pKa = 4.24SGDD109 pKa = 3.48NFVKK113 pKa = 10.5ISTSFKK119 pKa = 10.62VNNLNSSTYY128 pKa = 8.43NYY130 pKa = 10.97ASIVYY135 pKa = 10.15SYY137 pKa = 11.28LNPDD141 pKa = 3.05NFKK144 pKa = 10.91LVGINIHH151 pKa = 7.04DD152 pKa = 3.97NAVYY156 pKa = 10.43AIGFTMRR163 pKa = 11.84NGIEE167 pKa = 4.1SAEE170 pKa = 4.18PFWPGVPTNLTWSPGTTYY188 pKa = 11.36NLTMVKK194 pKa = 9.92QGSSLDD200 pKa = 3.76LLVNDD205 pKa = 4.9TKK207 pKa = 11.54YY208 pKa = 9.85LTQNNSDD215 pKa = 3.4NQTNFGDD222 pKa = 3.62VGVSYY227 pKa = 11.17GRR229 pKa = 11.84INNITFFDD237 pKa = 4.48FEE239 pKa = 4.73TEE241 pKa = 3.82ITDD244 pKa = 4.22QSVNSNNSQRR254 pKa = 11.84NTNPGPFTTSDD265 pKa = 3.16SQAILLEE272 pKa = 4.73GNSLPEE278 pKa = 3.67NEE280 pKa = 4.49YY281 pKa = 11.0LHH283 pKa = 7.31LYY285 pKa = 7.78DD286 pKa = 3.47TTPYY290 pKa = 9.9RR291 pKa = 11.84IEE293 pKa = 4.44SGHH296 pKa = 5.86VAAKK300 pKa = 9.93LPCGEE305 pKa = 4.97DD306 pKa = 3.43NMPEE310 pKa = 4.55AILLAGQAPTLAPIDD325 pKa = 3.87LEE327 pKa = 4.43FVNEE331 pKa = 3.9LSIPDD336 pKa = 4.01EE337 pKa = 4.2MCLYY341 pKa = 10.35HH342 pKa = 8.45ADD344 pKa = 3.77INSSNIDD351 pKa = 4.48PITDD355 pKa = 3.07IAIANNSTDD364 pKa = 4.12EE365 pKa = 3.95IDD367 pKa = 3.84FPEE370 pKa = 4.25TSGIVISISEE380 pKa = 3.81ISMIEE385 pKa = 4.5PIWMDD390 pKa = 3.57ASQTTLDD397 pKa = 3.53NN398 pKa = 4.0
MM1 pKa = 7.4SLHH4 pKa = 6.33NEE6 pKa = 3.69NMIKK10 pKa = 10.24EE11 pKa = 4.29FSLLLILGILIVTSSTSMSNPIDD34 pKa = 3.81AQLNSSDD41 pKa = 3.91TNIGNNGVMSIGSSSGFSNTNTTGMGIDD69 pKa = 5.87DD70 pKa = 4.01IDD72 pKa = 3.52SWDD75 pKa = 3.66VVSGNWINRR84 pKa = 11.84AFTIQGGLPEE94 pKa = 4.22NSGLNRR100 pKa = 11.84IILYY104 pKa = 9.76PEE106 pKa = 4.24SGDD109 pKa = 3.48NFVKK113 pKa = 10.5ISTSFKK119 pKa = 10.62VNNLNSSTYY128 pKa = 8.43NYY130 pKa = 10.97ASIVYY135 pKa = 10.15SYY137 pKa = 11.28LNPDD141 pKa = 3.05NFKK144 pKa = 10.91LVGINIHH151 pKa = 7.04DD152 pKa = 3.97NAVYY156 pKa = 10.43AIGFTMRR163 pKa = 11.84NGIEE167 pKa = 4.1SAEE170 pKa = 4.18PFWPGVPTNLTWSPGTTYY188 pKa = 11.36NLTMVKK194 pKa = 9.92QGSSLDD200 pKa = 3.76LLVNDD205 pKa = 4.9TKK207 pKa = 11.54YY208 pKa = 9.85LTQNNSDD215 pKa = 3.4NQTNFGDD222 pKa = 3.62VGVSYY227 pKa = 11.17GRR229 pKa = 11.84INNITFFDD237 pKa = 4.48FEE239 pKa = 4.73TEE241 pKa = 3.82ITDD244 pKa = 4.22QSVNSNNSQRR254 pKa = 11.84NTNPGPFTTSDD265 pKa = 3.16SQAILLEE272 pKa = 4.73GNSLPEE278 pKa = 3.67NEE280 pKa = 4.49YY281 pKa = 11.0LHH283 pKa = 7.31LYY285 pKa = 7.78DD286 pKa = 3.47TTPYY290 pKa = 9.9RR291 pKa = 11.84IEE293 pKa = 4.44SGHH296 pKa = 5.86VAAKK300 pKa = 9.93LPCGEE305 pKa = 4.97DD306 pKa = 3.43NMPEE310 pKa = 4.55AILLAGQAPTLAPIDD325 pKa = 3.87LEE327 pKa = 4.43FVNEE331 pKa = 3.9LSIPDD336 pKa = 4.01EE337 pKa = 4.2MCLYY341 pKa = 10.35HH342 pKa = 8.45ADD344 pKa = 3.77INSSNIDD351 pKa = 4.48PITDD355 pKa = 3.07IAIANNSTDD364 pKa = 4.12EE365 pKa = 3.95IDD367 pKa = 3.84FPEE370 pKa = 4.25TSGIVISISEE380 pKa = 3.81ISMIEE385 pKa = 4.5PIWMDD390 pKa = 3.57ASQTTLDD397 pKa = 3.53NN398 pKa = 4.0
Molecular weight: 43.46 kDa
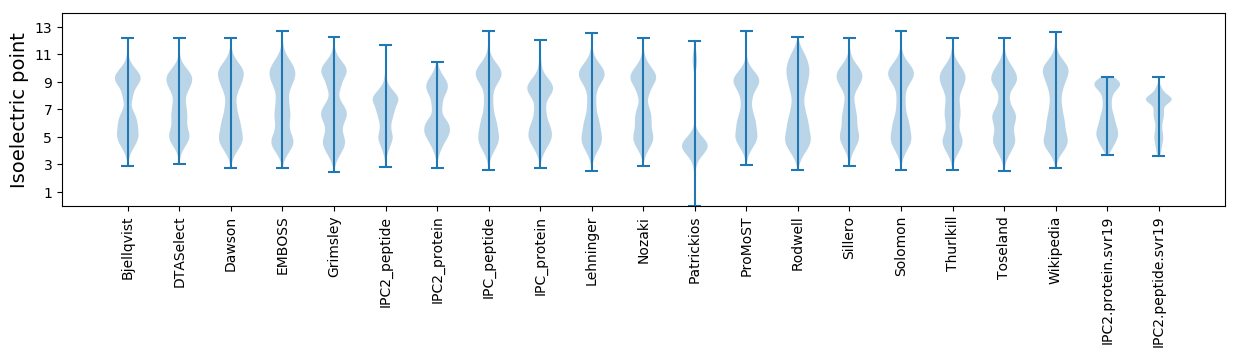
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A484I7N5|A0A484I7N5_9ARCH Uncharacterized protein OS=Candidatus Nitrosocosmicus franklandus OX=1798806 GN=NFRAN_0439 PE=4 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84CSKK6 pKa = 9.69CQKK9 pKa = 10.12GLHH12 pKa = 4.73TTYY15 pKa = 10.52IRR17 pKa = 11.84LGPKK21 pKa = 9.95AVFTKK26 pKa = 10.66LGYY29 pKa = 10.56YY30 pKa = 10.17CINCKK35 pKa = 10.38SYY37 pKa = 11.13TEE39 pKa = 4.37NNSQAFRR46 pKa = 11.84GARR49 pKa = 11.84KK50 pKa = 7.68QTQNRR55 pKa = 11.84MLRR58 pKa = 11.84PGFEE62 pKa = 4.16PGIVALRR69 pKa = 11.84GRR71 pKa = 11.84NAA73 pKa = 3.28
MM1 pKa = 7.39ARR3 pKa = 11.84CSKK6 pKa = 9.69CQKK9 pKa = 10.12GLHH12 pKa = 4.73TTYY15 pKa = 10.52IRR17 pKa = 11.84LGPKK21 pKa = 9.95AVFTKK26 pKa = 10.66LGYY29 pKa = 10.56YY30 pKa = 10.17CINCKK35 pKa = 10.38SYY37 pKa = 11.13TEE39 pKa = 4.37NNSQAFRR46 pKa = 11.84GARR49 pKa = 11.84KK50 pKa = 7.68QTQNRR55 pKa = 11.84MLRR58 pKa = 11.84PGFEE62 pKa = 4.16PGIVALRR69 pKa = 11.84GRR71 pKa = 11.84NAA73 pKa = 3.28
Molecular weight: 8.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
684365 |
32 |
1264 |
218.3 |
24.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.023 ± 0.05 | 1.035 ± 0.018 |
5.66 ± 0.037 | 6.155 ± 0.043 |
4.52 ± 0.039 | 6.026 ± 0.052 |
1.826 ± 0.022 | 9.435 ± 0.053 |
7.427 ± 0.052 | 9.223 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.431 ± 0.026 | 6.411 ± 0.045 |
3.633 ± 0.032 | 3.083 ± 0.033 |
3.998 ± 0.039 | 8.028 ± 0.048 |
5.482 ± 0.042 | 6.066 ± 0.04 |
0.888 ± 0.015 | 3.65 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
