
Sorex araneus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Alphapolyomavirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
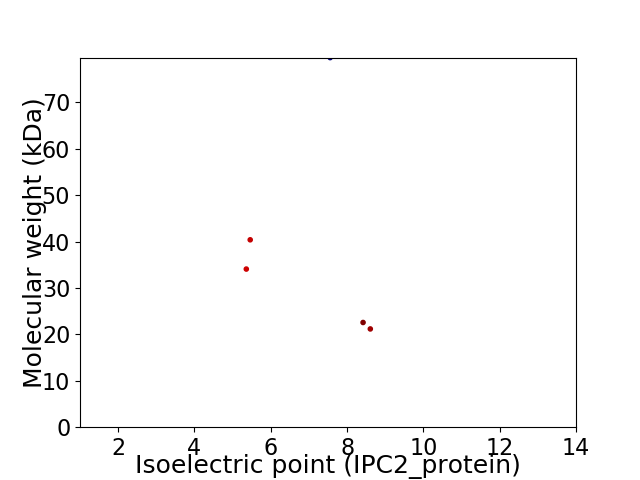
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223PYK3|A0A223PYK3_9POLY Minor capsid protein VP2 OS=Sorex araneus polyomavirus 1 OX=2560769 GN=VP3 PE=3 SV=1
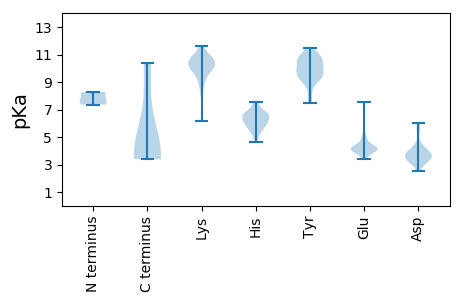
MM1 pKa = 7.37APKK4 pKa = 10.47RR5 pKa = 11.84KK6 pKa = 6.14TTCSSKK12 pKa = 9.45KK13 pKa = 8.44TCPQPSSVPKK23 pKa = 10.51LIIKK27 pKa = 10.25GGIEE31 pKa = 3.93VLDD34 pKa = 4.01VKK36 pKa = 10.35TGDD39 pKa = 4.0DD40 pKa = 4.31SITQIEE46 pKa = 4.54AFLNPRR52 pKa = 11.84MGVNDD57 pKa = 4.35EE58 pKa = 4.21TNTWYY63 pKa = 10.59GFSEE67 pKa = 4.38QVTVATARR75 pKa = 11.84EE76 pKa = 3.94TDD78 pKa = 3.64RR79 pKa = 11.84PPKK82 pKa = 10.11EE83 pKa = 3.54QMPYY87 pKa = 9.5YY88 pKa = 9.62SCARR92 pKa = 11.84IPLPLLNEE100 pKa = 4.49DD101 pKa = 3.74MTCNTLLMWEE111 pKa = 4.19AVSVKK116 pKa = 10.22TEE118 pKa = 4.18VIGSNTLMNVHH129 pKa = 7.52DD130 pKa = 4.39YY131 pKa = 7.47MTRR134 pKa = 11.84TDD136 pKa = 3.72NGVGHH141 pKa = 6.66PVVGSTYY148 pKa = 11.08HH149 pKa = 5.27MFAVGGEE156 pKa = 3.99PLDD159 pKa = 3.7LQGIQQSHH167 pKa = 6.0LVQYY171 pKa = 10.09PEE173 pKa = 3.97GLIVPKK179 pKa = 10.65SVTDD183 pKa = 3.39VTAKK187 pKa = 10.0IQCLDD192 pKa = 3.64PSAKK196 pKa = 10.4AKK198 pKa = 10.26LDD200 pKa = 3.43KK201 pKa = 10.76DD202 pKa = 3.46GKK204 pKa = 10.83YY205 pKa = 10.26PIEE208 pKa = 4.22TWSPDD213 pKa = 3.05PSRR216 pKa = 11.84NEE218 pKa = 3.42NTRR221 pKa = 11.84YY222 pKa = 9.46FGNYY226 pKa = 9.01YY227 pKa = 10.64GGLTTPPVLTFTNTVTTILLDD248 pKa = 3.88EE249 pKa = 5.0NGVGPLCKK257 pKa = 10.08GDD259 pKa = 4.6GLFLSCCDD267 pKa = 3.3VMGWFTAGSGTHH279 pKa = 4.67QRR281 pKa = 11.84FRR283 pKa = 11.84GLPRR287 pKa = 11.84YY288 pKa = 10.0FNVQLRR294 pKa = 11.84KK295 pKa = 9.21RR296 pKa = 11.84AVRR299 pKa = 11.84NPYY302 pKa = 9.14PVSALLTSLFTNMMPRR318 pKa = 11.84MSGQPMIGNKK328 pKa = 9.41SQVEE332 pKa = 4.1EE333 pKa = 4.0VRR335 pKa = 11.84VYY337 pKa = 10.85EE338 pKa = 4.78GLEE341 pKa = 3.9QLPGDD346 pKa = 4.14PDD348 pKa = 3.59MEE350 pKa = 4.08RR351 pKa = 11.84HH352 pKa = 5.77IDD354 pKa = 3.57EE355 pKa = 5.26FGQEE359 pKa = 4.0ITPVPP364 pKa = 3.72
MM1 pKa = 7.37APKK4 pKa = 10.47RR5 pKa = 11.84KK6 pKa = 6.14TTCSSKK12 pKa = 9.45KK13 pKa = 8.44TCPQPSSVPKK23 pKa = 10.51LIIKK27 pKa = 10.25GGIEE31 pKa = 3.93VLDD34 pKa = 4.01VKK36 pKa = 10.35TGDD39 pKa = 4.0DD40 pKa = 4.31SITQIEE46 pKa = 4.54AFLNPRR52 pKa = 11.84MGVNDD57 pKa = 4.35EE58 pKa = 4.21TNTWYY63 pKa = 10.59GFSEE67 pKa = 4.38QVTVATARR75 pKa = 11.84EE76 pKa = 3.94TDD78 pKa = 3.64RR79 pKa = 11.84PPKK82 pKa = 10.11EE83 pKa = 3.54QMPYY87 pKa = 9.5YY88 pKa = 9.62SCARR92 pKa = 11.84IPLPLLNEE100 pKa = 4.49DD101 pKa = 3.74MTCNTLLMWEE111 pKa = 4.19AVSVKK116 pKa = 10.22TEE118 pKa = 4.18VIGSNTLMNVHH129 pKa = 7.52DD130 pKa = 4.39YY131 pKa = 7.47MTRR134 pKa = 11.84TDD136 pKa = 3.72NGVGHH141 pKa = 6.66PVVGSTYY148 pKa = 11.08HH149 pKa = 5.27MFAVGGEE156 pKa = 3.99PLDD159 pKa = 3.7LQGIQQSHH167 pKa = 6.0LVQYY171 pKa = 10.09PEE173 pKa = 3.97GLIVPKK179 pKa = 10.65SVTDD183 pKa = 3.39VTAKK187 pKa = 10.0IQCLDD192 pKa = 3.64PSAKK196 pKa = 10.4AKK198 pKa = 10.26LDD200 pKa = 3.43KK201 pKa = 10.76DD202 pKa = 3.46GKK204 pKa = 10.83YY205 pKa = 10.26PIEE208 pKa = 4.22TWSPDD213 pKa = 3.05PSRR216 pKa = 11.84NEE218 pKa = 3.42NTRR221 pKa = 11.84YY222 pKa = 9.46FGNYY226 pKa = 9.01YY227 pKa = 10.64GGLTTPPVLTFTNTVTTILLDD248 pKa = 3.88EE249 pKa = 5.0NGVGPLCKK257 pKa = 10.08GDD259 pKa = 4.6GLFLSCCDD267 pKa = 3.3VMGWFTAGSGTHH279 pKa = 4.67QRR281 pKa = 11.84FRR283 pKa = 11.84GLPRR287 pKa = 11.84YY288 pKa = 10.0FNVQLRR294 pKa = 11.84KK295 pKa = 9.21RR296 pKa = 11.84AVRR299 pKa = 11.84NPYY302 pKa = 9.14PVSALLTSLFTNMMPRR318 pKa = 11.84MSGQPMIGNKK328 pKa = 9.41SQVEE332 pKa = 4.1EE333 pKa = 4.0VRR335 pKa = 11.84VYY337 pKa = 10.85EE338 pKa = 4.78GLEE341 pKa = 3.9QLPGDD346 pKa = 4.14PDD348 pKa = 3.59MEE350 pKa = 4.08RR351 pKa = 11.84HH352 pKa = 5.77IDD354 pKa = 3.57EE355 pKa = 5.26FGQEE359 pKa = 4.0ITPVPP364 pKa = 3.72
Molecular weight: 40.39 kDa
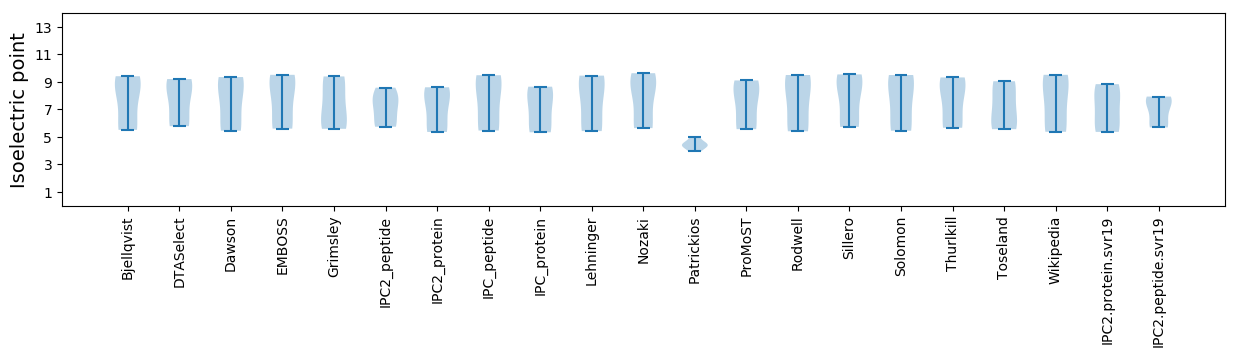
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223PYK8|A0A223PYK8_9POLY Large t antigen OS=Sorex araneus polyomavirus 1 OX=2560769 GN=LTAg PE=4 SV=1
MM1 pKa = 8.27DD2 pKa = 6.0SILTFAEE9 pKa = 4.02RR10 pKa = 11.84QLLISLLKK18 pKa = 10.52ISGDD22 pKa = 3.44TFGNVPAMARR32 pKa = 11.84AYY34 pKa = 9.7KK35 pKa = 10.24LAAKK39 pKa = 10.03RR40 pKa = 11.84LHH42 pKa = 6.69PDD44 pKa = 2.56KK45 pKa = 11.14GGNEE49 pKa = 3.97AEE51 pKa = 4.11MKK53 pKa = 10.37KK54 pKa = 10.82LNEE57 pKa = 3.92LWNKK61 pKa = 9.79FKK63 pKa = 11.22DD64 pKa = 3.97GIYY67 pKa = 10.41NLRR70 pKa = 11.84EE71 pKa = 4.3VKK73 pKa = 10.27PSLHH77 pKa = 6.26PVVTCTVLGARR88 pKa = 11.84NIFNLITNSSQCMRR102 pKa = 11.84NLLRR106 pKa = 11.84YY107 pKa = 8.49CRR109 pKa = 11.84CFCCILFQQHH119 pKa = 5.59RR120 pKa = 11.84QLKK123 pKa = 6.75ITYY126 pKa = 9.65RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84CNVWGQCYY137 pKa = 10.14CFLCYY142 pKa = 9.12YY143 pKa = 9.47TWFGVNCSIGAFTEE157 pKa = 3.93WLILLKK163 pKa = 10.81HH164 pKa = 6.36LDD166 pKa = 3.17WRR168 pKa = 11.84LLKK171 pKa = 10.39ISSAEE176 pKa = 3.79LDD178 pKa = 3.84VLGKK182 pKa = 10.4
MM1 pKa = 8.27DD2 pKa = 6.0SILTFAEE9 pKa = 4.02RR10 pKa = 11.84QLLISLLKK18 pKa = 10.52ISGDD22 pKa = 3.44TFGNVPAMARR32 pKa = 11.84AYY34 pKa = 9.7KK35 pKa = 10.24LAAKK39 pKa = 10.03RR40 pKa = 11.84LHH42 pKa = 6.69PDD44 pKa = 2.56KK45 pKa = 11.14GGNEE49 pKa = 3.97AEE51 pKa = 4.11MKK53 pKa = 10.37KK54 pKa = 10.82LNEE57 pKa = 3.92LWNKK61 pKa = 9.79FKK63 pKa = 11.22DD64 pKa = 3.97GIYY67 pKa = 10.41NLRR70 pKa = 11.84EE71 pKa = 4.3VKK73 pKa = 10.27PSLHH77 pKa = 6.26PVVTCTVLGARR88 pKa = 11.84NIFNLITNSSQCMRR102 pKa = 11.84NLLRR106 pKa = 11.84YY107 pKa = 8.49CRR109 pKa = 11.84CFCCILFQQHH119 pKa = 5.59RR120 pKa = 11.84QLKK123 pKa = 6.75ITYY126 pKa = 9.65RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84CNVWGQCYY137 pKa = 10.14CFLCYY142 pKa = 9.12YY143 pKa = 9.47TWFGVNCSIGAFTEE157 pKa = 3.93WLILLKK163 pKa = 10.81HH164 pKa = 6.36LDD166 pKa = 3.17WRR168 pKa = 11.84LLKK171 pKa = 10.39ISSAEE176 pKa = 3.79LDD178 pKa = 3.84VLGKK182 pKa = 10.4
Molecular weight: 21.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1764 |
182 |
708 |
352.8 |
39.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.519 ± 1.133 | 2.324 ± 0.739 |
5.102 ± 0.784 | 5.612 ± 0.355 |
4.025 ± 0.709 | 7.313 ± 0.796 |
2.494 ± 0.23 | 5.612 ± 0.376 |
6.066 ± 1.406 | 9.694 ± 1.137 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.685 ± 0.54 | 4.592 ± 0.826 |
5.612 ± 0.741 | 4.252 ± 0.661 |
5.215 ± 0.772 | 5.896 ± 0.594 |
5.839 ± 0.811 | 5.782 ± 0.643 |
1.587 ± 0.581 | 2.778 ± 0.322 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
