
Nile crocodilepox virus (isolate Crocodylus niloticus/Zimbabwe/Ume/2001) (CRV)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Chitovirales; Poxviridae; Chordopoxvirinae; Crocodylidpoxvirus; Nile crocodilepox virus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
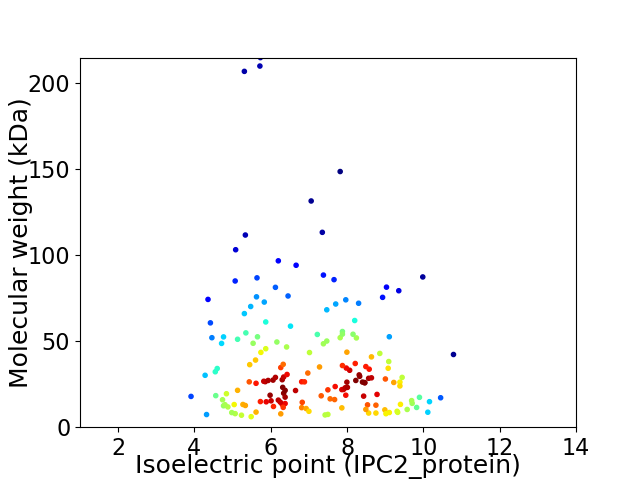
Virtual 2D-PAGE plot for 172 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q070N3|Q070N3_CPRVZ Uncharacterized protein OS=Nile crocodilepox virus (isolate Crocodylus niloticus/Zimbabwe/Ume/2001) OX=1289473 GN=CRV018 PE=4 SV=1
MM1 pKa = 7.59LEE3 pKa = 4.35AEE5 pKa = 4.82RR6 pKa = 11.84GDD8 pKa = 3.14AGYY11 pKa = 7.91FTDD14 pKa = 6.01GDD16 pKa = 4.22DD17 pKa = 4.43VDD19 pKa = 5.67DD20 pKa = 6.08GSDD23 pKa = 3.6PDD25 pKa = 3.85EE26 pKa = 6.15DD27 pKa = 3.96GAADD31 pKa = 3.92PFEE34 pKa = 5.11HH35 pKa = 7.19SIVEE39 pKa = 4.09YY40 pKa = 11.08DD41 pKa = 3.71RR42 pKa = 11.84FPPLSGIRR50 pKa = 11.84HH51 pKa = 6.74DD52 pKa = 4.63LLCVSITLGPEE63 pKa = 3.72TRR65 pKa = 11.84EE66 pKa = 3.67TLYY69 pKa = 11.11LRR71 pKa = 11.84FLNALGAVEE80 pKa = 4.52TIMRR84 pKa = 11.84VIPVSALEE92 pKa = 4.0EE93 pKa = 4.07PFAYY97 pKa = 9.67EE98 pKa = 3.78HH99 pKa = 6.42QIVLSRR105 pKa = 11.84EE106 pKa = 3.91YY107 pKa = 10.3FSRR110 pKa = 11.84YY111 pKa = 9.4AAMDD115 pKa = 3.34MMACSSPDD123 pKa = 3.33APDD126 pKa = 3.57PCAYY130 pKa = 9.42IFSAAGIDD138 pKa = 3.97AINAFTASARR148 pKa = 11.84DD149 pKa = 3.78PEE151 pKa = 4.42EE152 pKa = 4.59EE153 pKa = 4.02PTATYY158 pKa = 10.92QFFVV162 pKa = 3.57
MM1 pKa = 7.59LEE3 pKa = 4.35AEE5 pKa = 4.82RR6 pKa = 11.84GDD8 pKa = 3.14AGYY11 pKa = 7.91FTDD14 pKa = 6.01GDD16 pKa = 4.22DD17 pKa = 4.43VDD19 pKa = 5.67DD20 pKa = 6.08GSDD23 pKa = 3.6PDD25 pKa = 3.85EE26 pKa = 6.15DD27 pKa = 3.96GAADD31 pKa = 3.92PFEE34 pKa = 5.11HH35 pKa = 7.19SIVEE39 pKa = 4.09YY40 pKa = 11.08DD41 pKa = 3.71RR42 pKa = 11.84FPPLSGIRR50 pKa = 11.84HH51 pKa = 6.74DD52 pKa = 4.63LLCVSITLGPEE63 pKa = 3.72TRR65 pKa = 11.84EE66 pKa = 3.67TLYY69 pKa = 11.11LRR71 pKa = 11.84FLNALGAVEE80 pKa = 4.52TIMRR84 pKa = 11.84VIPVSALEE92 pKa = 4.0EE93 pKa = 4.07PFAYY97 pKa = 9.67EE98 pKa = 3.78HH99 pKa = 6.42QIVLSRR105 pKa = 11.84EE106 pKa = 3.91YY107 pKa = 10.3FSRR110 pKa = 11.84YY111 pKa = 9.4AAMDD115 pKa = 3.34MMACSSPDD123 pKa = 3.33APDD126 pKa = 3.57PCAYY130 pKa = 9.42IFSAAGIDD138 pKa = 3.97AINAFTASARR148 pKa = 11.84DD149 pKa = 3.78PEE151 pKa = 4.42EE152 pKa = 4.59EE153 pKa = 4.02PTATYY158 pKa = 10.92QFFVV162 pKa = 3.57
Molecular weight: 17.88 kDa
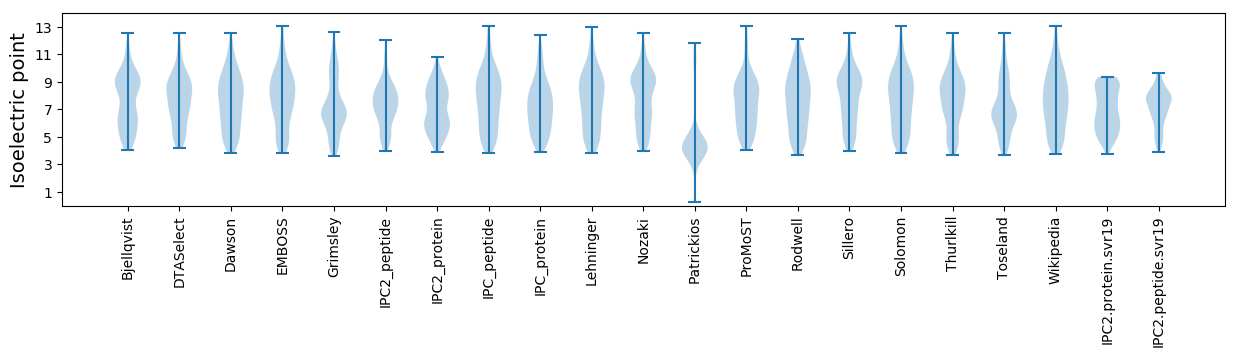
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q06ZY5|Q06ZY5_CPRVZ Uncharacterized protein OS=Nile crocodilepox virus (isolate Crocodylus niloticus/Zimbabwe/Ume/2001) OX=1289473 GN=CRV166 PE=4 SV=1
MM1 pKa = 7.33RR2 pKa = 11.84THH4 pKa = 7.2LFLSVFLRR12 pKa = 11.84FLLPRR17 pKa = 11.84RR18 pKa = 11.84PSRR21 pKa = 11.84ATSCRR26 pKa = 11.84AAAPRR31 pKa = 11.84CAPRR35 pKa = 11.84SRR37 pKa = 11.84PRR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84PPRR45 pKa = 11.84PGPPRR50 pKa = 11.84PGSRR54 pKa = 11.84SRR56 pKa = 11.84ACGCRR61 pKa = 11.84GRR63 pKa = 11.84RR64 pKa = 11.84ARR66 pKa = 11.84LLRR69 pKa = 11.84LPLPLLPRR77 pKa = 11.84ARR79 pKa = 11.84SRR81 pKa = 11.84LPRR84 pKa = 11.84PRR86 pKa = 11.84RR87 pKa = 11.84PPLAAPALARR97 pKa = 11.84LPRR100 pKa = 11.84LPPRR104 pKa = 11.84ARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84SRR113 pKa = 11.84PRR115 pKa = 11.84PRR117 pKa = 11.84DD118 pKa = 2.96RR119 pKa = 11.84PAARR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84APPRR129 pKa = 11.84TRR131 pKa = 11.84LPPPPPRR138 pKa = 11.84CRR140 pKa = 11.84PSPSRR145 pKa = 11.84SPPSRR150 pKa = 11.84RR151 pKa = 11.84SRR153 pKa = 11.84PRR155 pKa = 11.84AASSSPSTPTPPPSSPPLPTRR176 pKa = 11.84PPPSPRR182 pKa = 11.84PRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84PRR188 pKa = 11.84AATATGFAPVLALVLDD204 pKa = 4.31PAPRR208 pKa = 11.84SSATRR213 pKa = 11.84SEE215 pKa = 4.18EE216 pKa = 3.93VPRR219 pKa = 11.84SGRR222 pKa = 11.84LRR224 pKa = 11.84RR225 pKa = 11.84TAFSRR230 pKa = 11.84SLLLFFLKK238 pKa = 10.41PPPFARR244 pKa = 11.84LASLASVPPLAPLARR259 pKa = 11.84DD260 pKa = 3.86LVARR264 pKa = 11.84SAPPPSAFPSSLAGKK279 pKa = 9.17GNRR282 pKa = 11.84GEE284 pKa = 4.27ARR286 pKa = 11.84ATASRR291 pKa = 11.84AFASVRR297 pKa = 11.84PASLARR303 pKa = 11.84RR304 pKa = 11.84LLPPPSRR311 pKa = 11.84PRR313 pKa = 11.84SRR315 pKa = 11.84EE316 pKa = 3.39KK317 pKa = 10.1GTEE320 pKa = 3.8EE321 pKa = 3.67RR322 pKa = 11.84RR323 pKa = 11.84ARR325 pKa = 11.84PPPAPSPPFARR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84SLGASSLRR346 pKa = 11.84LPVLARR352 pKa = 11.84GKK354 pKa = 9.78RR355 pKa = 11.84EE356 pKa = 3.47PRR358 pKa = 11.84RR359 pKa = 11.84GVRR362 pKa = 11.84CSRR365 pKa = 11.84ARR367 pKa = 11.84NDD369 pKa = 2.92AAARR373 pKa = 11.84RR374 pKa = 11.84TKK376 pKa = 10.54KK377 pKa = 10.5KK378 pKa = 10.15EE379 pKa = 3.71EE380 pKa = 4.02KK381 pKa = 10.61
MM1 pKa = 7.33RR2 pKa = 11.84THH4 pKa = 7.2LFLSVFLRR12 pKa = 11.84FLLPRR17 pKa = 11.84RR18 pKa = 11.84PSRR21 pKa = 11.84ATSCRR26 pKa = 11.84AAAPRR31 pKa = 11.84CAPRR35 pKa = 11.84SRR37 pKa = 11.84PRR39 pKa = 11.84SRR41 pKa = 11.84RR42 pKa = 11.84PPRR45 pKa = 11.84PGPPRR50 pKa = 11.84PGSRR54 pKa = 11.84SRR56 pKa = 11.84ACGCRR61 pKa = 11.84GRR63 pKa = 11.84RR64 pKa = 11.84ARR66 pKa = 11.84LLRR69 pKa = 11.84LPLPLLPRR77 pKa = 11.84ARR79 pKa = 11.84SRR81 pKa = 11.84LPRR84 pKa = 11.84PRR86 pKa = 11.84RR87 pKa = 11.84PPLAAPALARR97 pKa = 11.84LPRR100 pKa = 11.84LPPRR104 pKa = 11.84ARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84RR111 pKa = 11.84SRR113 pKa = 11.84PRR115 pKa = 11.84PRR117 pKa = 11.84DD118 pKa = 2.96RR119 pKa = 11.84PAARR123 pKa = 11.84RR124 pKa = 11.84RR125 pKa = 11.84APPRR129 pKa = 11.84TRR131 pKa = 11.84LPPPPPRR138 pKa = 11.84CRR140 pKa = 11.84PSPSRR145 pKa = 11.84SPPSRR150 pKa = 11.84RR151 pKa = 11.84SRR153 pKa = 11.84PRR155 pKa = 11.84AASSSPSTPTPPPSSPPLPTRR176 pKa = 11.84PPPSPRR182 pKa = 11.84PRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84PRR188 pKa = 11.84AATATGFAPVLALVLDD204 pKa = 4.31PAPRR208 pKa = 11.84SSATRR213 pKa = 11.84SEE215 pKa = 4.18EE216 pKa = 3.93VPRR219 pKa = 11.84SGRR222 pKa = 11.84LRR224 pKa = 11.84RR225 pKa = 11.84TAFSRR230 pKa = 11.84SLLLFFLKK238 pKa = 10.41PPPFARR244 pKa = 11.84LASLASVPPLAPLARR259 pKa = 11.84DD260 pKa = 3.86LVARR264 pKa = 11.84SAPPPSAFPSSLAGKK279 pKa = 9.17GNRR282 pKa = 11.84GEE284 pKa = 4.27ARR286 pKa = 11.84ATASRR291 pKa = 11.84AFASVRR297 pKa = 11.84PASLARR303 pKa = 11.84RR304 pKa = 11.84LLPPPSRR311 pKa = 11.84PRR313 pKa = 11.84SRR315 pKa = 11.84EE316 pKa = 3.39KK317 pKa = 10.1GTEE320 pKa = 3.8EE321 pKa = 3.67RR322 pKa = 11.84RR323 pKa = 11.84ARR325 pKa = 11.84PPPAPSPPFARR336 pKa = 11.84RR337 pKa = 11.84RR338 pKa = 11.84SLGASSLRR346 pKa = 11.84LPVLARR352 pKa = 11.84GKK354 pKa = 9.78RR355 pKa = 11.84EE356 pKa = 3.47PRR358 pKa = 11.84RR359 pKa = 11.84GVRR362 pKa = 11.84CSRR365 pKa = 11.84ARR367 pKa = 11.84NDD369 pKa = 2.92AAARR373 pKa = 11.84RR374 pKa = 11.84TKK376 pKa = 10.54KK377 pKa = 10.5KK378 pKa = 10.15EE379 pKa = 3.71EE380 pKa = 4.02KK381 pKa = 10.61
Molecular weight: 42.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
59641 |
53 |
1941 |
346.8 |
38.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.92 ± 0.41 | 2.113 ± 0.088 |
6.195 ± 0.133 | 5.761 ± 0.125 |
4.604 ± 0.148 | 5.795 ± 0.245 |
1.856 ± 0.078 | 4.489 ± 0.189 |
3.711 ± 0.191 | 9.74 ± 0.199 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.88 ± 0.066 | 3.944 ± 0.154 |
5.508 ± 0.281 | 2.264 ± 0.102 |
8.56 ± 0.291 | 7.567 ± 0.216 |
4.755 ± 0.112 | 7.741 ± 0.161 |
0.676 ± 0.036 | 3.923 ± 0.129 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
