
Sewage-associated circular DNA virus-1
Taxonomy: Viruses; unclassified viruses
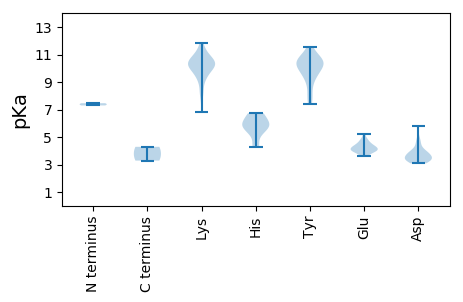
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
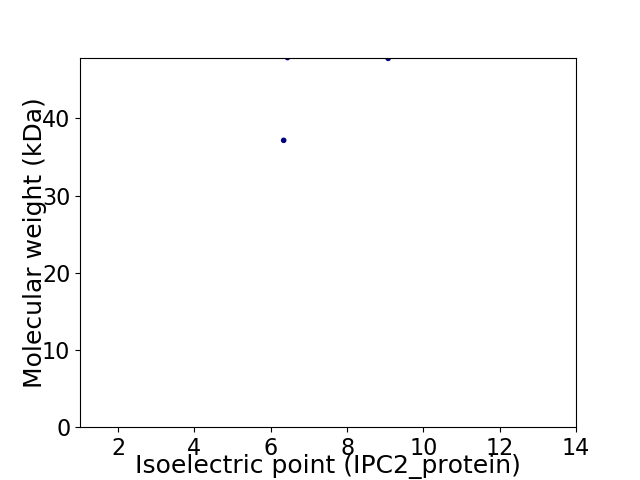
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
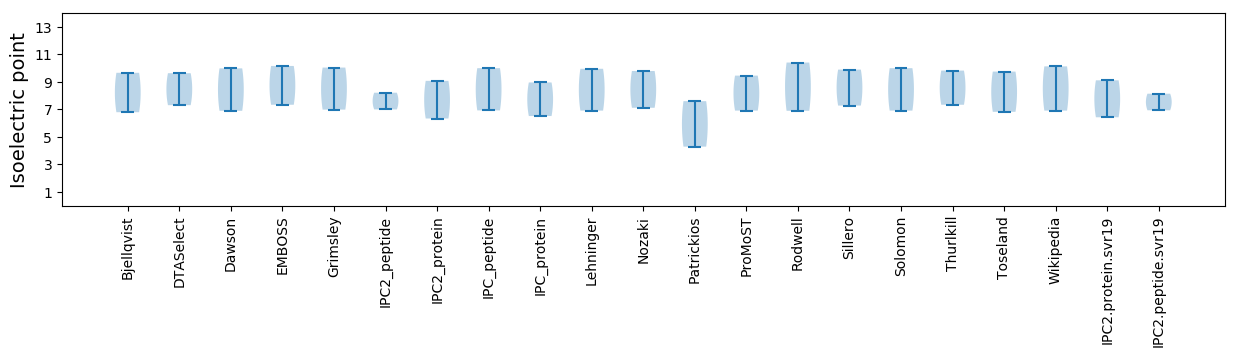
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075J166|A0A075J166_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-1 OX=1519385 PE=3 SV=1
MM1 pKa = 7.3PPAWEE6 pKa = 4.56GSSFIITYY14 pKa = 7.81PQSDD18 pKa = 4.0FDD20 pKa = 3.95LQEE23 pKa = 5.24YY24 pKa = 10.05IDD26 pKa = 3.29TWEE29 pKa = 4.04NKK31 pKa = 8.53KK32 pKa = 10.17HH33 pKa = 5.99IKK35 pKa = 9.9YY36 pKa = 10.6LLISTEE42 pKa = 3.93LHH44 pKa = 5.61QDD46 pKa = 3.26EE47 pKa = 4.91SLHH50 pKa = 5.4RR51 pKa = 11.84HH52 pKa = 5.19AVLHH56 pKa = 6.02FSKK59 pKa = 10.33KK60 pKa = 7.89QRR62 pKa = 11.84VGHH65 pKa = 6.56AFFDD69 pKa = 3.57HH70 pKa = 6.73LTRR73 pKa = 11.84HH74 pKa = 6.03PNIKK78 pKa = 10.2CVGKK82 pKa = 10.49RR83 pKa = 11.84RR84 pKa = 11.84SDD86 pKa = 3.24WTNVIAYY93 pKa = 7.95VKK95 pKa = 10.28KK96 pKa = 10.48CGEE99 pKa = 3.95FVEE102 pKa = 4.64WGEE105 pKa = 4.23PRR107 pKa = 11.84HH108 pKa = 5.81SACVWSAIATSSTRR122 pKa = 11.84QEE124 pKa = 4.04AQEE127 pKa = 3.91MLSIEE132 pKa = 4.32RR133 pKa = 11.84PRR135 pKa = 11.84DD136 pKa = 3.2AVLNARR142 pKa = 11.84NFDD145 pKa = 3.38YY146 pKa = 10.71WLDD149 pKa = 3.49KK150 pKa = 10.3VFPMSEE156 pKa = 3.92TASYY160 pKa = 10.29SGRR163 pKa = 11.84SSDD166 pKa = 3.73QFVVPPALQDD176 pKa = 3.34WVLMSWWYY184 pKa = 9.9VLPRR188 pKa = 11.84QEE190 pKa = 4.18RR191 pKa = 11.84PKK193 pKa = 11.15SLVLNGASRR202 pKa = 11.84LGKK205 pKa = 6.84TQWARR210 pKa = 11.84SLGPHH215 pKa = 6.66IYY217 pKa = 9.57IGNMWDD223 pKa = 3.87LGALDD228 pKa = 5.84GIDD231 pKa = 3.68SNFWNYY237 pKa = 10.37GYY239 pKa = 10.34IVFDD243 pKa = 4.93DD244 pKa = 4.53IEE246 pKa = 4.2WASIAGSAKK255 pKa = 10.01SWFGAQRR262 pKa = 11.84DD263 pKa = 3.88FSVSDD268 pKa = 3.18KK269 pKa = 10.74YY270 pKa = 11.01KK271 pKa = 10.71RR272 pKa = 11.84RR273 pKa = 11.84TGWWYY278 pKa = 8.65VTSTNEE284 pKa = 3.66NGDD287 pKa = 3.64WLVVFLAFFLSTLTLIAVIVIVLLTQNGDD316 pKa = 3.32VKK318 pKa = 10.25TLII321 pKa = 4.29
MM1 pKa = 7.3PPAWEE6 pKa = 4.56GSSFIITYY14 pKa = 7.81PQSDD18 pKa = 4.0FDD20 pKa = 3.95LQEE23 pKa = 5.24YY24 pKa = 10.05IDD26 pKa = 3.29TWEE29 pKa = 4.04NKK31 pKa = 8.53KK32 pKa = 10.17HH33 pKa = 5.99IKK35 pKa = 9.9YY36 pKa = 10.6LLISTEE42 pKa = 3.93LHH44 pKa = 5.61QDD46 pKa = 3.26EE47 pKa = 4.91SLHH50 pKa = 5.4RR51 pKa = 11.84HH52 pKa = 5.19AVLHH56 pKa = 6.02FSKK59 pKa = 10.33KK60 pKa = 7.89QRR62 pKa = 11.84VGHH65 pKa = 6.56AFFDD69 pKa = 3.57HH70 pKa = 6.73LTRR73 pKa = 11.84HH74 pKa = 6.03PNIKK78 pKa = 10.2CVGKK82 pKa = 10.49RR83 pKa = 11.84RR84 pKa = 11.84SDD86 pKa = 3.24WTNVIAYY93 pKa = 7.95VKK95 pKa = 10.28KK96 pKa = 10.48CGEE99 pKa = 3.95FVEE102 pKa = 4.64WGEE105 pKa = 4.23PRR107 pKa = 11.84HH108 pKa = 5.81SACVWSAIATSSTRR122 pKa = 11.84QEE124 pKa = 4.04AQEE127 pKa = 3.91MLSIEE132 pKa = 4.32RR133 pKa = 11.84PRR135 pKa = 11.84DD136 pKa = 3.2AVLNARR142 pKa = 11.84NFDD145 pKa = 3.38YY146 pKa = 10.71WLDD149 pKa = 3.49KK150 pKa = 10.3VFPMSEE156 pKa = 3.92TASYY160 pKa = 10.29SGRR163 pKa = 11.84SSDD166 pKa = 3.73QFVVPPALQDD176 pKa = 3.34WVLMSWWYY184 pKa = 9.9VLPRR188 pKa = 11.84QEE190 pKa = 4.18RR191 pKa = 11.84PKK193 pKa = 11.15SLVLNGASRR202 pKa = 11.84LGKK205 pKa = 6.84TQWARR210 pKa = 11.84SLGPHH215 pKa = 6.66IYY217 pKa = 9.57IGNMWDD223 pKa = 3.87LGALDD228 pKa = 5.84GIDD231 pKa = 3.68SNFWNYY237 pKa = 10.37GYY239 pKa = 10.34IVFDD243 pKa = 4.93DD244 pKa = 4.53IEE246 pKa = 4.2WASIAGSAKK255 pKa = 10.01SWFGAQRR262 pKa = 11.84DD263 pKa = 3.88FSVSDD268 pKa = 3.18KK269 pKa = 10.74YY270 pKa = 11.01KK271 pKa = 10.71RR272 pKa = 11.84RR273 pKa = 11.84TGWWYY278 pKa = 8.65VTSTNEE284 pKa = 3.66NGDD287 pKa = 3.64WLVVFLAFFLSTLTLIAVIVIVLLTQNGDD316 pKa = 3.32VKK318 pKa = 10.25TLII321 pKa = 4.29
Molecular weight: 37.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075J166|A0A075J166_9VIRU Replication-associated protein OS=Sewage-associated circular DNA virus-1 OX=1519385 PE=3 SV=1
MM1 pKa = 7.46SKK3 pKa = 11.22YY4 pKa = 7.4MTPKK8 pKa = 10.14NVARR12 pKa = 11.84VINFAQQAKK21 pKa = 8.4RR22 pKa = 11.84TWDD25 pKa = 3.53SPGMRR30 pKa = 11.84TVRR33 pKa = 11.84AAGKK37 pKa = 9.58AARR40 pKa = 11.84SRR42 pKa = 11.84IAANRR47 pKa = 11.84LRR49 pKa = 11.84GKK51 pKa = 9.75KK52 pKa = 9.87SKK54 pKa = 10.77GFGGPSSGHH63 pKa = 4.26YY64 pKa = 9.72VGPFKK69 pKa = 10.74RR70 pKa = 11.84PKK72 pKa = 9.21PVKK75 pKa = 9.49KK76 pKa = 10.58NKK78 pKa = 9.54IADD81 pKa = 3.72YY82 pKa = 10.42LSQGVCEE89 pKa = 4.18YY90 pKa = 11.23KK91 pKa = 10.39EE92 pKa = 3.59IHH94 pKa = 6.66GNVSDD99 pKa = 4.73PDD101 pKa = 3.63CVYY104 pKa = 10.77ATFSTWDD111 pKa = 3.27DD112 pKa = 3.86SMVEE116 pKa = 3.98RR117 pKa = 11.84QLYY120 pKa = 9.44GALLRR125 pKa = 11.84KK126 pKa = 9.22LLKK129 pKa = 10.37KK130 pKa = 10.68AGINIDD136 pKa = 4.12RR137 pKa = 11.84NDD139 pKa = 3.29QVLPMSQYY147 pKa = 10.06NNSGGTLIQAVFKK160 pKa = 10.52SVSGSVLIYY169 pKa = 10.72EE170 pKa = 4.85DD171 pKa = 4.45LLGGAQSIQSIVSQDD186 pKa = 3.91LIKK189 pKa = 10.94VGLWFKK195 pKa = 11.56VMVKK199 pKa = 10.1GQQSLTFPDD208 pKa = 3.09QSYY211 pKa = 10.23QLEE214 pKa = 4.29SLSMYY219 pKa = 9.52GQSTTHH225 pKa = 5.61SAEE228 pKa = 4.1RR229 pKa = 11.84HH230 pKa = 5.88CLAQLNVMQEE240 pKa = 3.95VLQIEE245 pKa = 4.58MFLEE249 pKa = 3.94ISIQNRR255 pKa = 11.84TKK257 pKa = 10.88GAIAGTGDD265 pKa = 3.53TEE267 pKa = 4.19TDD269 pKa = 3.4RR270 pKa = 11.84VDD272 pKa = 3.5SQPLQGKK279 pKa = 9.62LYY281 pKa = 8.91EE282 pKa = 4.47FNGGVPKK289 pKa = 10.28EE290 pKa = 3.99RR291 pKa = 11.84QKK293 pKa = 11.85GNGLLTEE300 pKa = 4.27VEE302 pKa = 4.2ALGISLNRR310 pKa = 11.84AAQIGNEE317 pKa = 4.3YY318 pKa = 10.51KK319 pKa = 10.42EE320 pKa = 4.44PPPAKK325 pKa = 10.04HH326 pKa = 6.02FSNVSKK332 pKa = 10.84VANQKK337 pKa = 9.05LQPGSIKK344 pKa = 10.11RR345 pKa = 11.84HH346 pKa = 4.37YY347 pKa = 9.46LKK349 pKa = 9.83WYY351 pKa = 10.46ARR353 pKa = 11.84GFVNNLVNRR362 pKa = 11.84WSSKK366 pKa = 8.1QNPTFGQTVVASPGKK381 pKa = 9.19CAMFAFEE388 pKa = 4.76EE389 pKa = 4.44VLNSGSSNLITVSYY403 pKa = 10.38EE404 pKa = 3.59VDD406 pKa = 3.35KK407 pKa = 11.56KK408 pKa = 11.15CGAKK412 pKa = 10.54LITAKK417 pKa = 10.28AAPMKK422 pKa = 10.05PLYY425 pKa = 10.46TSDD428 pKa = 3.97TYY430 pKa = 11.56SSPP433 pKa = 3.29
MM1 pKa = 7.46SKK3 pKa = 11.22YY4 pKa = 7.4MTPKK8 pKa = 10.14NVARR12 pKa = 11.84VINFAQQAKK21 pKa = 8.4RR22 pKa = 11.84TWDD25 pKa = 3.53SPGMRR30 pKa = 11.84TVRR33 pKa = 11.84AAGKK37 pKa = 9.58AARR40 pKa = 11.84SRR42 pKa = 11.84IAANRR47 pKa = 11.84LRR49 pKa = 11.84GKK51 pKa = 9.75KK52 pKa = 9.87SKK54 pKa = 10.77GFGGPSSGHH63 pKa = 4.26YY64 pKa = 9.72VGPFKK69 pKa = 10.74RR70 pKa = 11.84PKK72 pKa = 9.21PVKK75 pKa = 9.49KK76 pKa = 10.58NKK78 pKa = 9.54IADD81 pKa = 3.72YY82 pKa = 10.42LSQGVCEE89 pKa = 4.18YY90 pKa = 11.23KK91 pKa = 10.39EE92 pKa = 3.59IHH94 pKa = 6.66GNVSDD99 pKa = 4.73PDD101 pKa = 3.63CVYY104 pKa = 10.77ATFSTWDD111 pKa = 3.27DD112 pKa = 3.86SMVEE116 pKa = 3.98RR117 pKa = 11.84QLYY120 pKa = 9.44GALLRR125 pKa = 11.84KK126 pKa = 9.22LLKK129 pKa = 10.37KK130 pKa = 10.68AGINIDD136 pKa = 4.12RR137 pKa = 11.84NDD139 pKa = 3.29QVLPMSQYY147 pKa = 10.06NNSGGTLIQAVFKK160 pKa = 10.52SVSGSVLIYY169 pKa = 10.72EE170 pKa = 4.85DD171 pKa = 4.45LLGGAQSIQSIVSQDD186 pKa = 3.91LIKK189 pKa = 10.94VGLWFKK195 pKa = 11.56VMVKK199 pKa = 10.1GQQSLTFPDD208 pKa = 3.09QSYY211 pKa = 10.23QLEE214 pKa = 4.29SLSMYY219 pKa = 9.52GQSTTHH225 pKa = 5.61SAEE228 pKa = 4.1RR229 pKa = 11.84HH230 pKa = 5.88CLAQLNVMQEE240 pKa = 3.95VLQIEE245 pKa = 4.58MFLEE249 pKa = 3.94ISIQNRR255 pKa = 11.84TKK257 pKa = 10.88GAIAGTGDD265 pKa = 3.53TEE267 pKa = 4.19TDD269 pKa = 3.4RR270 pKa = 11.84VDD272 pKa = 3.5SQPLQGKK279 pKa = 9.62LYY281 pKa = 8.91EE282 pKa = 4.47FNGGVPKK289 pKa = 10.28EE290 pKa = 3.99RR291 pKa = 11.84QKK293 pKa = 11.85GNGLLTEE300 pKa = 4.27VEE302 pKa = 4.2ALGISLNRR310 pKa = 11.84AAQIGNEE317 pKa = 4.3YY318 pKa = 10.51KK319 pKa = 10.42EE320 pKa = 4.44PPPAKK325 pKa = 10.04HH326 pKa = 6.02FSNVSKK332 pKa = 10.84VANQKK337 pKa = 9.05LQPGSIKK344 pKa = 10.11RR345 pKa = 11.84HH346 pKa = 4.37YY347 pKa = 9.46LKK349 pKa = 9.83WYY351 pKa = 10.46ARR353 pKa = 11.84GFVNNLVNRR362 pKa = 11.84WSSKK366 pKa = 8.1QNPTFGQTVVASPGKK381 pKa = 9.19CAMFAFEE388 pKa = 4.76EE389 pKa = 4.44VLNSGSSNLITVSYY403 pKa = 10.38EE404 pKa = 3.59VDD406 pKa = 3.35KK407 pKa = 11.56KK408 pKa = 11.15CGAKK412 pKa = 10.54LITAKK417 pKa = 10.28AAPMKK422 pKa = 10.05PLYY425 pKa = 10.46TSDD428 pKa = 3.97TYY430 pKa = 11.56SSPP433 pKa = 3.29
Molecular weight: 47.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
754 |
321 |
433 |
377.0 |
42.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.029 ± 0.296 | 1.061 ± 0.077 |
4.775 ± 0.886 | 4.509 ± 0.1 |
3.979 ± 0.612 | 7.162 ± 0.946 |
2.122 ± 0.604 | 5.172 ± 0.454 |
7.029 ± 1.244 | 8.09 ± 0.195 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.122 ± 0.343 | 4.642 ± 0.55 |
4.244 ± 0.308 | 5.172 ± 0.872 |
5.04 ± 0.345 | 9.151 ± 0.071 |
4.775 ± 0.128 | 7.162 ± 0.002 |
2.918 ± 1.447 | 3.846 ± 0.066 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
