
Capybara microvirus Cap3_SP_443
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.4
Get precalculated fractions of proteins
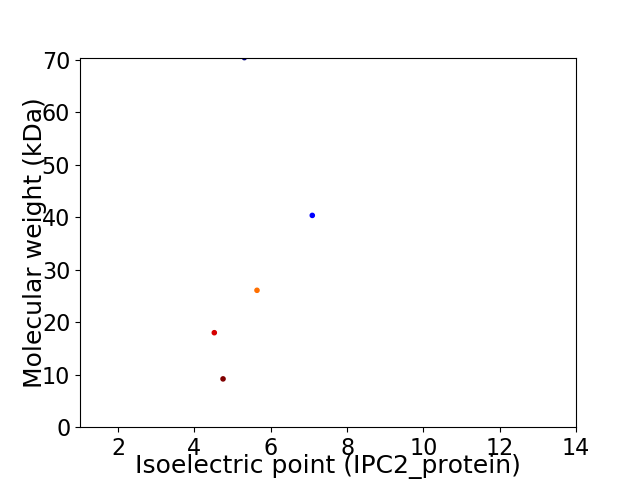
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
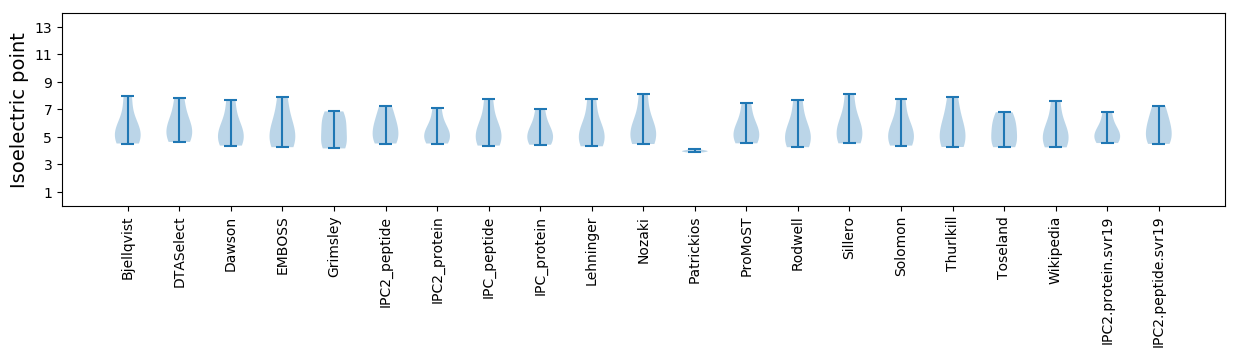
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8L3|A0A4P8W8L3_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_443 OX=2585455 PE=4 SV=1
MM1 pKa = 7.29KK2 pKa = 10.54VKK4 pKa = 10.21IYY6 pKa = 10.58SAYY9 pKa = 10.36DD10 pKa = 3.38LPKK13 pKa = 10.15PSEE16 pKa = 4.06LTGFSPIEE24 pKa = 3.83KK25 pKa = 9.42TYY27 pKa = 10.11MEE29 pKa = 4.87QYY31 pKa = 10.61NKK33 pKa = 10.92DD34 pKa = 3.53GIKK37 pKa = 10.21EE38 pKa = 3.98IIEE41 pKa = 4.01VGEE44 pKa = 4.39TNWYY48 pKa = 10.9AEE50 pKa = 4.29VQSYY54 pKa = 11.56ADD56 pKa = 3.49EE57 pKa = 4.4TDD59 pKa = 3.14VNKK62 pKa = 10.43IMARR66 pKa = 11.84VAGGEE71 pKa = 4.13TGLLDD76 pKa = 3.75QMKK79 pKa = 10.4GFYY82 pKa = 10.4LDD84 pKa = 3.75TQGLPASLMEE94 pKa = 4.37AQNIILKK101 pKa = 8.67GQKK104 pKa = 10.45AFDD107 pKa = 3.91NLPAEE112 pKa = 4.43IRR114 pKa = 11.84RR115 pKa = 11.84MYY117 pKa = 11.22DD118 pKa = 2.78NDD120 pKa = 3.21AMKK123 pKa = 10.67FMANFDD129 pKa = 3.71YY130 pKa = 11.31DD131 pKa = 3.74QVAKK135 pKa = 10.77NLEE138 pKa = 4.01EE139 pKa = 4.2VQAGKK144 pKa = 10.52AVTQAVKK151 pKa = 10.74EE152 pKa = 4.23GGNDD156 pKa = 3.22NAEE159 pKa = 4.2NAA161 pKa = 4.07
MM1 pKa = 7.29KK2 pKa = 10.54VKK4 pKa = 10.21IYY6 pKa = 10.58SAYY9 pKa = 10.36DD10 pKa = 3.38LPKK13 pKa = 10.15PSEE16 pKa = 4.06LTGFSPIEE24 pKa = 3.83KK25 pKa = 9.42TYY27 pKa = 10.11MEE29 pKa = 4.87QYY31 pKa = 10.61NKK33 pKa = 10.92DD34 pKa = 3.53GIKK37 pKa = 10.21EE38 pKa = 3.98IIEE41 pKa = 4.01VGEE44 pKa = 4.39TNWYY48 pKa = 10.9AEE50 pKa = 4.29VQSYY54 pKa = 11.56ADD56 pKa = 3.49EE57 pKa = 4.4TDD59 pKa = 3.14VNKK62 pKa = 10.43IMARR66 pKa = 11.84VAGGEE71 pKa = 4.13TGLLDD76 pKa = 3.75QMKK79 pKa = 10.4GFYY82 pKa = 10.4LDD84 pKa = 3.75TQGLPASLMEE94 pKa = 4.37AQNIILKK101 pKa = 8.67GQKK104 pKa = 10.45AFDD107 pKa = 3.91NLPAEE112 pKa = 4.43IRR114 pKa = 11.84RR115 pKa = 11.84MYY117 pKa = 11.22DD118 pKa = 2.78NDD120 pKa = 3.21AMKK123 pKa = 10.67FMANFDD129 pKa = 3.71YY130 pKa = 11.31DD131 pKa = 3.74QVAKK135 pKa = 10.77NLEE138 pKa = 4.01EE139 pKa = 4.2VQAGKK144 pKa = 10.52AVTQAVKK151 pKa = 10.74EE152 pKa = 4.23GGNDD156 pKa = 3.22NAEE159 pKa = 4.2NAA161 pKa = 4.07
Molecular weight: 18.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8L3|A0A4P8W8L3_9VIRU Replication initiator protein OS=Capybara microvirus Cap3_SP_443 OX=2585455 PE=4 SV=1
MM1 pKa = 7.74ACQKK5 pKa = 9.24PLCFIEE11 pKa = 4.27YY12 pKa = 9.04PGEE15 pKa = 3.84RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 9.95ILPARR23 pKa = 11.84IWDD26 pKa = 3.81EE27 pKa = 3.72YY28 pKa = 11.12LAAKK32 pKa = 10.11RR33 pKa = 11.84DD34 pKa = 3.7GDD36 pKa = 3.94EE37 pKa = 4.33EE38 pKa = 4.44TLSRR42 pKa = 11.84APYY45 pKa = 8.63YY46 pKa = 10.44GATRR50 pKa = 11.84VVQIPCGQCIQCRR63 pKa = 11.84LDD65 pKa = 4.9KK66 pKa = 10.67SRR68 pKa = 11.84KK69 pKa = 6.06WANRR73 pKa = 11.84MMAEE77 pKa = 4.16ARR79 pKa = 11.84CWDD82 pKa = 3.31HH83 pKa = 7.45NYY85 pKa = 9.79FVTLTYY91 pKa = 10.94DD92 pKa = 3.87DD93 pKa = 3.68NHH95 pKa = 7.74LPIRR99 pKa = 11.84DD100 pKa = 3.62WFHH103 pKa = 7.93DD104 pKa = 3.77EE105 pKa = 4.94DD106 pKa = 5.17GSIQEE111 pKa = 4.81GVTLLPDD118 pKa = 4.77EE119 pKa = 4.83ISKK122 pKa = 10.18FMKK125 pKa = 10.22KK126 pKa = 10.07LRR128 pKa = 11.84VYY130 pKa = 9.38YY131 pKa = 9.85EE132 pKa = 3.71RR133 pKa = 11.84RR134 pKa = 11.84TGKK137 pKa = 8.51TGIRR141 pKa = 11.84FFGCGEE147 pKa = 3.95YY148 pKa = 10.83GEE150 pKa = 4.75KK151 pKa = 9.91SGRR154 pKa = 11.84PHH156 pKa = 4.84YY157 pKa = 10.48HH158 pKa = 6.7IILFNCEE165 pKa = 4.09LDD167 pKa = 3.59DD168 pKa = 4.64LKK170 pKa = 10.88PYY172 pKa = 10.34KK173 pKa = 9.93ATEE176 pKa = 4.25CGPLFNSAFLEE187 pKa = 4.74SIWSDD192 pKa = 3.3EE193 pKa = 4.18YY194 pKa = 10.84GQKK197 pKa = 10.86GYY199 pKa = 8.54VTVAPFTWDD208 pKa = 2.51TGAYY212 pKa = 7.04VARR215 pKa = 11.84YY216 pKa = 9.23IMKK219 pKa = 9.96KK220 pKa = 9.54QLGPPAYY227 pKa = 9.91SYY229 pKa = 11.58YY230 pKa = 10.36NEE232 pKa = 3.93HH233 pKa = 6.65RR234 pKa = 11.84KK235 pKa = 9.4IPEE238 pKa = 3.7YY239 pKa = 11.1VIMSRR244 pKa = 11.84NPGIGKK250 pKa = 9.79FYY252 pKa = 10.78YY253 pKa = 9.88DD254 pKa = 3.32QKK256 pKa = 11.11KK257 pKa = 10.88DD258 pKa = 3.64EE259 pKa = 4.57IYY261 pKa = 9.26EE262 pKa = 3.84TDD264 pKa = 3.42EE265 pKa = 5.05IIIHH269 pKa = 5.62KK270 pKa = 10.23HH271 pKa = 4.3GATIKK276 pKa = 10.35AKK278 pKa = 9.39PGKK281 pKa = 10.49YY282 pKa = 9.67FDD284 pKa = 4.18GYY286 pKa = 10.09YY287 pKa = 10.46AAEE290 pKa = 4.39EE291 pKa = 4.29PEE293 pKa = 3.95KK294 pKa = 10.82MEE296 pKa = 4.07VIKK299 pKa = 10.59RR300 pKa = 11.84KK301 pKa = 9.66RR302 pKa = 11.84EE303 pKa = 3.55RR304 pKa = 11.84AGYY307 pKa = 9.08MSEE310 pKa = 4.19LEE312 pKa = 3.98QRR314 pKa = 11.84RR315 pKa = 11.84QSGLTAQEE323 pKa = 3.97YY324 pKa = 10.42RR325 pKa = 11.84DD326 pKa = 3.8LQAEE330 pKa = 4.29SMARR334 pKa = 11.84RR335 pKa = 11.84MAGLSRR341 pKa = 11.84DD342 pKa = 3.38LDD344 pKa = 3.72
MM1 pKa = 7.74ACQKK5 pKa = 9.24PLCFIEE11 pKa = 4.27YY12 pKa = 9.04PGEE15 pKa = 3.84RR16 pKa = 11.84RR17 pKa = 11.84KK18 pKa = 9.95ILPARR23 pKa = 11.84IWDD26 pKa = 3.81EE27 pKa = 3.72YY28 pKa = 11.12LAAKK32 pKa = 10.11RR33 pKa = 11.84DD34 pKa = 3.7GDD36 pKa = 3.94EE37 pKa = 4.33EE38 pKa = 4.44TLSRR42 pKa = 11.84APYY45 pKa = 8.63YY46 pKa = 10.44GATRR50 pKa = 11.84VVQIPCGQCIQCRR63 pKa = 11.84LDD65 pKa = 4.9KK66 pKa = 10.67SRR68 pKa = 11.84KK69 pKa = 6.06WANRR73 pKa = 11.84MMAEE77 pKa = 4.16ARR79 pKa = 11.84CWDD82 pKa = 3.31HH83 pKa = 7.45NYY85 pKa = 9.79FVTLTYY91 pKa = 10.94DD92 pKa = 3.87DD93 pKa = 3.68NHH95 pKa = 7.74LPIRR99 pKa = 11.84DD100 pKa = 3.62WFHH103 pKa = 7.93DD104 pKa = 3.77EE105 pKa = 4.94DD106 pKa = 5.17GSIQEE111 pKa = 4.81GVTLLPDD118 pKa = 4.77EE119 pKa = 4.83ISKK122 pKa = 10.18FMKK125 pKa = 10.22KK126 pKa = 10.07LRR128 pKa = 11.84VYY130 pKa = 9.38YY131 pKa = 9.85EE132 pKa = 3.71RR133 pKa = 11.84RR134 pKa = 11.84TGKK137 pKa = 8.51TGIRR141 pKa = 11.84FFGCGEE147 pKa = 3.95YY148 pKa = 10.83GEE150 pKa = 4.75KK151 pKa = 9.91SGRR154 pKa = 11.84PHH156 pKa = 4.84YY157 pKa = 10.48HH158 pKa = 6.7IILFNCEE165 pKa = 4.09LDD167 pKa = 3.59DD168 pKa = 4.64LKK170 pKa = 10.88PYY172 pKa = 10.34KK173 pKa = 9.93ATEE176 pKa = 4.25CGPLFNSAFLEE187 pKa = 4.74SIWSDD192 pKa = 3.3EE193 pKa = 4.18YY194 pKa = 10.84GQKK197 pKa = 10.86GYY199 pKa = 8.54VTVAPFTWDD208 pKa = 2.51TGAYY212 pKa = 7.04VARR215 pKa = 11.84YY216 pKa = 9.23IMKK219 pKa = 9.96KK220 pKa = 9.54QLGPPAYY227 pKa = 9.91SYY229 pKa = 11.58YY230 pKa = 10.36NEE232 pKa = 3.93HH233 pKa = 6.65RR234 pKa = 11.84KK235 pKa = 9.4IPEE238 pKa = 3.7YY239 pKa = 11.1VIMSRR244 pKa = 11.84NPGIGKK250 pKa = 9.79FYY252 pKa = 10.78YY253 pKa = 9.88DD254 pKa = 3.32QKK256 pKa = 11.11KK257 pKa = 10.88DD258 pKa = 3.64EE259 pKa = 4.57IYY261 pKa = 9.26EE262 pKa = 3.84TDD264 pKa = 3.42EE265 pKa = 5.05IIIHH269 pKa = 5.62KK270 pKa = 10.23HH271 pKa = 4.3GATIKK276 pKa = 10.35AKK278 pKa = 9.39PGKK281 pKa = 10.49YY282 pKa = 9.67FDD284 pKa = 4.18GYY286 pKa = 10.09YY287 pKa = 10.46AAEE290 pKa = 4.39EE291 pKa = 4.29PEE293 pKa = 3.95KK294 pKa = 10.82MEE296 pKa = 4.07VIKK299 pKa = 10.59RR300 pKa = 11.84KK301 pKa = 9.66RR302 pKa = 11.84EE303 pKa = 3.55RR304 pKa = 11.84AGYY307 pKa = 9.08MSEE310 pKa = 4.19LEE312 pKa = 3.98QRR314 pKa = 11.84RR315 pKa = 11.84QSGLTAQEE323 pKa = 3.97YY324 pKa = 10.42RR325 pKa = 11.84DD326 pKa = 3.8LQAEE330 pKa = 4.29SMARR334 pKa = 11.84RR335 pKa = 11.84MAGLSRR341 pKa = 11.84DD342 pKa = 3.38LDD344 pKa = 3.72
Molecular weight: 40.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1466 |
79 |
625 |
293.2 |
32.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.458 ± 2.108 | 0.75 ± 0.476 |
5.866 ± 0.822 | 7.094 ± 0.643 |
3.888 ± 0.744 | 8.595 ± 0.738 |
1.842 ± 0.287 | 4.911 ± 0.597 |
6.071 ± 0.642 | 6.003 ± 0.804 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.411 ± 0.267 | 4.638 ± 0.827 |
4.025 ± 0.822 | 4.57 ± 0.339 |
4.707 ± 0.927 | 7.094 ± 1.646 |
5.662 ± 0.695 | 5.457 ± 0.732 |
1.569 ± 0.29 | 5.389 ± 0.647 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
