
Streptococcus phage Javan414
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
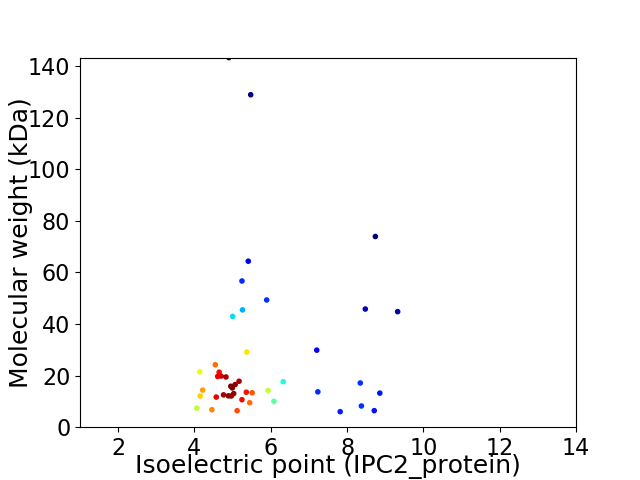
Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6B6Q6|A0A4D6B6Q6_9CAUD Capsid and scaffold protein OS=Streptococcus phage Javan414 OX=2548155 GN=Javan414_0005 PE=4 SV=1
MM1 pKa = 7.38KK2 pKa = 10.1KK3 pKa = 10.19KK4 pKa = 10.79NEE6 pKa = 3.93GAVRR10 pKa = 11.84NLIALCLITLAVVLGIAAFGAVVKK34 pKa = 10.25EE35 pKa = 4.03KK36 pKa = 10.62QSEE39 pKa = 4.26NKK41 pKa = 9.69IVTSSSSNVDD51 pKa = 3.36DD52 pKa = 4.51SDD54 pKa = 5.04DD55 pKa = 3.84YY56 pKa = 11.98SSEE59 pKa = 4.11DD60 pKa = 3.69DD61 pKa = 3.31NDD63 pKa = 3.73YY64 pKa = 11.29NYY66 pKa = 11.19SSSSSSNYY74 pKa = 9.56SSSSSSEE81 pKa = 3.39IFDD84 pKa = 4.12PNNYY88 pKa = 8.41EE89 pKa = 4.19VPDD92 pKa = 4.14FATWNHH98 pKa = 6.36DD99 pKa = 3.55QLEE102 pKa = 4.41HH103 pKa = 6.93DD104 pKa = 4.86KK105 pKa = 10.79KK106 pKa = 10.72IQITGTVVQNMEE118 pKa = 4.53DD119 pKa = 3.35DD120 pKa = 4.58GIYY123 pKa = 9.71YY124 pKa = 10.34LRR126 pKa = 11.84VAMDD130 pKa = 4.22DD131 pKa = 4.49DD132 pKa = 4.25YY133 pKa = 12.07DD134 pKa = 5.58KK135 pKa = 11.27IVMVGISKK143 pKa = 10.24YY144 pKa = 11.04VYY146 pKa = 10.19DD147 pKa = 5.18DD148 pKa = 4.88VIAEE152 pKa = 4.25DD153 pKa = 4.82DD154 pKa = 3.66NVTFYY159 pKa = 11.37GVAKK163 pKa = 10.45GLTSYY168 pKa = 11.09EE169 pKa = 4.08STLGKK174 pKa = 8.53TVTLPLMYY182 pKa = 10.42GHH184 pKa = 7.09YY185 pKa = 10.85YY186 pKa = 10.3NINSYY191 pKa = 11.39GNN193 pKa = 3.34
MM1 pKa = 7.38KK2 pKa = 10.1KK3 pKa = 10.19KK4 pKa = 10.79NEE6 pKa = 3.93GAVRR10 pKa = 11.84NLIALCLITLAVVLGIAAFGAVVKK34 pKa = 10.25EE35 pKa = 4.03KK36 pKa = 10.62QSEE39 pKa = 4.26NKK41 pKa = 9.69IVTSSSSNVDD51 pKa = 3.36DD52 pKa = 4.51SDD54 pKa = 5.04DD55 pKa = 3.84YY56 pKa = 11.98SSEE59 pKa = 4.11DD60 pKa = 3.69DD61 pKa = 3.31NDD63 pKa = 3.73YY64 pKa = 11.29NYY66 pKa = 11.19SSSSSSNYY74 pKa = 9.56SSSSSSEE81 pKa = 3.39IFDD84 pKa = 4.12PNNYY88 pKa = 8.41EE89 pKa = 4.19VPDD92 pKa = 4.14FATWNHH98 pKa = 6.36DD99 pKa = 3.55QLEE102 pKa = 4.41HH103 pKa = 6.93DD104 pKa = 4.86KK105 pKa = 10.79KK106 pKa = 10.72IQITGTVVQNMEE118 pKa = 4.53DD119 pKa = 3.35DD120 pKa = 4.58GIYY123 pKa = 9.71YY124 pKa = 10.34LRR126 pKa = 11.84VAMDD130 pKa = 4.22DD131 pKa = 4.49DD132 pKa = 4.25YY133 pKa = 12.07DD134 pKa = 5.58KK135 pKa = 11.27IVMVGISKK143 pKa = 10.24YY144 pKa = 11.04VYY146 pKa = 10.19DD147 pKa = 5.18DD148 pKa = 4.88VIAEE152 pKa = 4.25DD153 pKa = 4.82DD154 pKa = 3.66NVTFYY159 pKa = 11.37GVAKK163 pKa = 10.45GLTSYY168 pKa = 11.09EE169 pKa = 4.08STLGKK174 pKa = 8.53TVTLPLMYY182 pKa = 10.42GHH184 pKa = 7.09YY185 pKa = 10.85YY186 pKa = 10.3NINSYY191 pKa = 11.39GNN193 pKa = 3.34
Molecular weight: 21.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6B6V3|A0A4D6B6V3_9CAUD Uncharacterized protein OS=Streptococcus phage Javan414 OX=2548155 GN=Javan414_0039 PE=4 SV=1
MM1 pKa = 7.37KK2 pKa = 10.17RR3 pKa = 11.84NKK5 pKa = 8.81TKK7 pKa = 10.32YY8 pKa = 9.53PSIFTYY14 pKa = 7.28EE15 pKa = 4.01TKK17 pKa = 10.15KK18 pKa = 10.43GKK20 pKa = 9.9RR21 pKa = 11.84YY22 pKa = 8.44YY23 pKa = 10.58VRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.71FKK29 pKa = 10.87LHH31 pKa = 6.28GKK33 pKa = 6.06QTEE36 pKa = 4.03ASASGLKK43 pKa = 9.01TLAEE47 pKa = 4.14ARR49 pKa = 11.84QALAEE54 pKa = 4.15IEE56 pKa = 4.27NKK58 pKa = 9.36IANGDD63 pKa = 3.46YY64 pKa = 10.76DD65 pKa = 3.63PRR67 pKa = 11.84KK68 pKa = 10.68NMTVDD73 pKa = 4.89DD74 pKa = 3.9YY75 pKa = 11.29WQIYY79 pKa = 8.63SDD81 pKa = 3.58NRR83 pKa = 11.84IKK85 pKa = 9.93TGRR88 pKa = 11.84WAPDD92 pKa = 3.78TIATKK97 pKa = 9.77MSWYY101 pKa = 9.83KK102 pKa = 10.36YY103 pKa = 9.51HH104 pKa = 6.28FHH106 pKa = 6.35KK107 pKa = 10.95QFGRR111 pKa = 11.84TLLKK115 pKa = 10.41DD116 pKa = 3.57IKK118 pKa = 8.48RR119 pKa = 11.84TKK121 pKa = 9.6YY122 pKa = 8.42EE123 pKa = 3.99AYY125 pKa = 9.88VSSMLKK131 pKa = 9.97NYY133 pKa = 10.31SRR135 pKa = 11.84TSVIQISALFEE146 pKa = 4.44SMLTDD151 pKa = 4.23AVINDD156 pKa = 4.49YY157 pKa = 10.76IDD159 pKa = 3.63KK160 pKa = 10.99NPILRR165 pKa = 11.84IYY167 pKa = 9.63IGKK170 pKa = 9.76SDD172 pKa = 3.65IPAKK176 pKa = 10.27KK177 pKa = 9.74KK178 pKa = 10.44RR179 pKa = 11.84LSLEE183 pKa = 4.32DD184 pKa = 3.46FRR186 pKa = 11.84IWDD189 pKa = 3.36KK190 pKa = 10.74CARR193 pKa = 11.84EE194 pKa = 4.01ILSSYY199 pKa = 10.11DD200 pKa = 3.22YY201 pKa = 11.51TMVRR205 pKa = 11.84LTYY208 pKa = 9.91FGLRR212 pKa = 11.84RR213 pKa = 11.84SEE215 pKa = 3.92VLGVKK220 pKa = 10.42FGSLSLVDD228 pKa = 3.7GRR230 pKa = 11.84FKK232 pKa = 10.84IFLDD236 pKa = 3.56EE237 pKa = 4.3SRR239 pKa = 11.84TSRR242 pKa = 11.84RR243 pKa = 11.84PEE245 pKa = 3.81GGRR248 pKa = 11.84MKK250 pKa = 10.4TKK252 pKa = 9.25TSEE255 pKa = 3.98RR256 pKa = 11.84YY257 pKa = 9.47VLVDD261 pKa = 3.39EE262 pKa = 4.84EE263 pKa = 4.68TTEE266 pKa = 4.24LLQKK270 pKa = 11.28AMILSRR276 pKa = 11.84QIAKK280 pKa = 7.27EE281 pKa = 3.93TNRR284 pKa = 11.84ILGQGDD290 pKa = 4.81FIFVDD295 pKa = 3.37TGMKK299 pKa = 9.74IKK301 pKa = 10.11NHH303 pKa = 6.29KK304 pKa = 9.37GNPIGYY310 pKa = 8.16SHH312 pKa = 7.66IAYY315 pKa = 9.9AFRR318 pKa = 11.84KK319 pKa = 10.27VSDD322 pKa = 3.24KK323 pKa = 10.96TNIYY327 pKa = 7.62VTPHH331 pKa = 4.66TMRR334 pKa = 11.84HH335 pKa = 5.08FFATQGQIAGVPVEE349 pKa = 4.8HH350 pKa = 6.66MAAALGHH357 pKa = 5.31STSYY361 pKa = 7.77MTQKK365 pKa = 8.95YY366 pKa = 6.41THH368 pKa = 6.9IKK370 pKa = 10.67DD371 pKa = 3.64EE372 pKa = 4.38VASEE376 pKa = 4.27VTDD379 pKa = 3.47SFLRR383 pKa = 11.84AIKK386 pKa = 10.4
MM1 pKa = 7.37KK2 pKa = 10.17RR3 pKa = 11.84NKK5 pKa = 8.81TKK7 pKa = 10.32YY8 pKa = 9.53PSIFTYY14 pKa = 7.28EE15 pKa = 4.01TKK17 pKa = 10.15KK18 pKa = 10.43GKK20 pKa = 9.9RR21 pKa = 11.84YY22 pKa = 8.44YY23 pKa = 10.58VRR25 pKa = 11.84RR26 pKa = 11.84KK27 pKa = 9.71FKK29 pKa = 10.87LHH31 pKa = 6.28GKK33 pKa = 6.06QTEE36 pKa = 4.03ASASGLKK43 pKa = 9.01TLAEE47 pKa = 4.14ARR49 pKa = 11.84QALAEE54 pKa = 4.15IEE56 pKa = 4.27NKK58 pKa = 9.36IANGDD63 pKa = 3.46YY64 pKa = 10.76DD65 pKa = 3.63PRR67 pKa = 11.84KK68 pKa = 10.68NMTVDD73 pKa = 4.89DD74 pKa = 3.9YY75 pKa = 11.29WQIYY79 pKa = 8.63SDD81 pKa = 3.58NRR83 pKa = 11.84IKK85 pKa = 9.93TGRR88 pKa = 11.84WAPDD92 pKa = 3.78TIATKK97 pKa = 9.77MSWYY101 pKa = 9.83KK102 pKa = 10.36YY103 pKa = 9.51HH104 pKa = 6.28FHH106 pKa = 6.35KK107 pKa = 10.95QFGRR111 pKa = 11.84TLLKK115 pKa = 10.41DD116 pKa = 3.57IKK118 pKa = 8.48RR119 pKa = 11.84TKK121 pKa = 9.6YY122 pKa = 8.42EE123 pKa = 3.99AYY125 pKa = 9.88VSSMLKK131 pKa = 9.97NYY133 pKa = 10.31SRR135 pKa = 11.84TSVIQISALFEE146 pKa = 4.44SMLTDD151 pKa = 4.23AVINDD156 pKa = 4.49YY157 pKa = 10.76IDD159 pKa = 3.63KK160 pKa = 10.99NPILRR165 pKa = 11.84IYY167 pKa = 9.63IGKK170 pKa = 9.76SDD172 pKa = 3.65IPAKK176 pKa = 10.27KK177 pKa = 9.74KK178 pKa = 10.44RR179 pKa = 11.84LSLEE183 pKa = 4.32DD184 pKa = 3.46FRR186 pKa = 11.84IWDD189 pKa = 3.36KK190 pKa = 10.74CARR193 pKa = 11.84EE194 pKa = 4.01ILSSYY199 pKa = 10.11DD200 pKa = 3.22YY201 pKa = 11.51TMVRR205 pKa = 11.84LTYY208 pKa = 9.91FGLRR212 pKa = 11.84RR213 pKa = 11.84SEE215 pKa = 3.92VLGVKK220 pKa = 10.42FGSLSLVDD228 pKa = 3.7GRR230 pKa = 11.84FKK232 pKa = 10.84IFLDD236 pKa = 3.56EE237 pKa = 4.3SRR239 pKa = 11.84TSRR242 pKa = 11.84RR243 pKa = 11.84PEE245 pKa = 3.81GGRR248 pKa = 11.84MKK250 pKa = 10.4TKK252 pKa = 9.25TSEE255 pKa = 3.98RR256 pKa = 11.84YY257 pKa = 9.47VLVDD261 pKa = 3.39EE262 pKa = 4.84EE263 pKa = 4.68TTEE266 pKa = 4.24LLQKK270 pKa = 11.28AMILSRR276 pKa = 11.84QIAKK280 pKa = 7.27EE281 pKa = 3.93TNRR284 pKa = 11.84ILGQGDD290 pKa = 4.81FIFVDD295 pKa = 3.37TGMKK299 pKa = 9.74IKK301 pKa = 10.11NHH303 pKa = 6.29KK304 pKa = 9.37GNPIGYY310 pKa = 8.16SHH312 pKa = 7.66IAYY315 pKa = 9.9AFRR318 pKa = 11.84KK319 pKa = 10.27VSDD322 pKa = 3.24KK323 pKa = 10.96TNIYY327 pKa = 7.62VTPHH331 pKa = 4.66TMRR334 pKa = 11.84HH335 pKa = 5.08FFATQGQIAGVPVEE349 pKa = 4.8HH350 pKa = 6.66MAAALGHH357 pKa = 5.31STSYY361 pKa = 7.77MTQKK365 pKa = 8.95YY366 pKa = 6.41THH368 pKa = 6.9IKK370 pKa = 10.67DD371 pKa = 3.64EE372 pKa = 4.38VASEE376 pKa = 4.27VTDD379 pKa = 3.47SFLRR383 pKa = 11.84AIKK386 pKa = 10.4
Molecular weight: 44.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10746 |
52 |
1347 |
238.8 |
26.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.314 ± 0.698 | 0.614 ± 0.128 |
6.57 ± 0.299 | 7.51 ± 0.511 |
4.029 ± 0.312 | 6.635 ± 0.34 |
1.126 ± 0.128 | 6.458 ± 0.341 |
8.217 ± 0.374 | 7.854 ± 0.349 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.227 | 5.667 ± 0.264 |
2.671 ± 0.142 | 3.732 ± 0.308 |
3.797 ± 0.289 | 6.309 ± 0.406 |
7.398 ± 0.577 | 6.235 ± 0.251 |
1.405 ± 0.18 | 3.992 ± 0.372 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
