
Lake Sinai virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Magsaviricetes; Nodamuvirales; Sinhaliviridae; Sinaivirus
Average proteome isoelectric point is 7.81
Get precalculated fractions of proteins
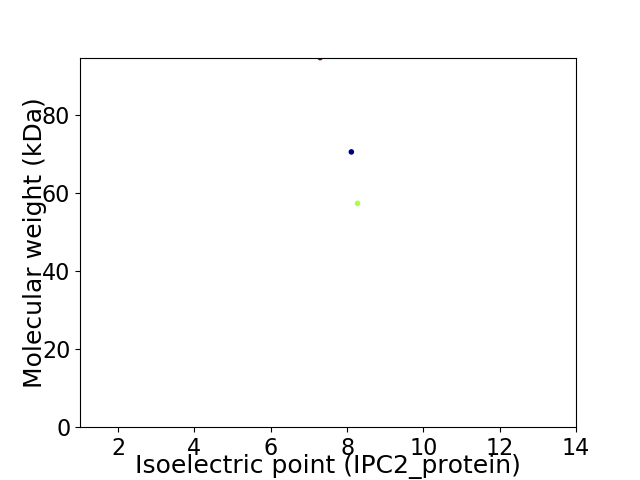
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A223DNF2|A0A223DNF2_9VIRU Capsid protein OS=Lake Sinai virus 2 OX=1041831 PE=4 SV=1
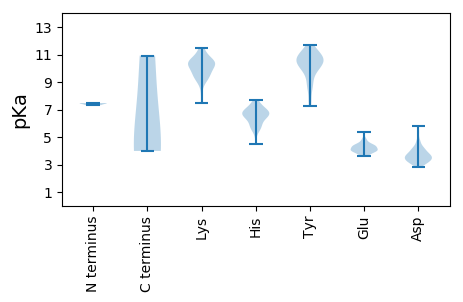
MM1 pKa = 7.32KK2 pKa = 10.52VVFLVSFAVFLICQVPPVSYY22 pKa = 9.91WLEE25 pKa = 4.26DD26 pKa = 3.85LLDD29 pKa = 4.76EE30 pKa = 4.59YY31 pKa = 11.57NTMCSYY37 pKa = 10.91EE38 pKa = 4.02YY39 pKa = 10.97AAAEE43 pKa = 4.11AYY45 pKa = 10.86NNFVHH50 pKa = 6.58KK51 pKa = 10.34HH52 pKa = 4.47RR53 pKa = 11.84VAAYY57 pKa = 8.32AAGVRR62 pKa = 11.84ILRR65 pKa = 11.84YY66 pKa = 7.27RR67 pKa = 11.84TPWYY71 pKa = 10.01CFASRR76 pKa = 11.84SVPVVHH82 pKa = 6.76PFSWLMGQYY91 pKa = 10.43DD92 pKa = 3.72ATIDD96 pKa = 3.47RR97 pKa = 11.84VGTLMQRR104 pKa = 11.84LEE106 pKa = 4.02IANVSISDD114 pKa = 4.68GIRR117 pKa = 11.84TVADD121 pKa = 3.65TALKK125 pKa = 10.44AYY127 pKa = 9.85FYY129 pKa = 10.87YY130 pKa = 10.4QIIWFQLFSIIMLVGFFSAVICSSRR155 pKa = 11.84VRR157 pKa = 11.84VVRR160 pKa = 11.84IRR162 pKa = 11.84NRR164 pKa = 11.84VRR166 pKa = 11.84GYY168 pKa = 10.63SVTEE172 pKa = 3.71LRR174 pKa = 11.84SDD176 pKa = 3.94FEE178 pKa = 4.49EE179 pKa = 4.56SMSAILAPLSRR190 pKa = 11.84GHH192 pKa = 6.3SCLNFQRR199 pKa = 11.84RR200 pKa = 11.84VVEE203 pKa = 4.36SWAIDD208 pKa = 3.18QLLRR212 pKa = 11.84YY213 pKa = 7.7FRR215 pKa = 11.84CFRR218 pKa = 11.84SVASSQGRR226 pKa = 11.84WAEE229 pKa = 3.97VGHH232 pKa = 6.34RR233 pKa = 11.84LHH235 pKa = 6.79RR236 pKa = 11.84CSPVVLDD243 pKa = 3.81GAFVPEE249 pKa = 3.7YY250 pKa = 10.1DD251 pKa = 4.07QRR253 pKa = 11.84FSACRR258 pKa = 11.84RR259 pKa = 11.84HH260 pKa = 6.64PSVCPDD266 pKa = 3.32RR267 pKa = 11.84FDD269 pKa = 3.29IPAAILSHH277 pKa = 6.53VDD279 pKa = 3.35YY280 pKa = 11.57YY281 pKa = 8.69MTPDD285 pKa = 3.22QLAAAVTGPTFIVNHH300 pKa = 7.26DD301 pKa = 3.8YY302 pKa = 11.66SSIDD306 pKa = 3.43TLSVAEE312 pKa = 4.05VSLRR316 pKa = 11.84SAGGLVTASVRR327 pKa = 11.84DD328 pKa = 3.95GPTFGPHH335 pKa = 7.18PYY337 pKa = 9.92YY338 pKa = 10.24HH339 pKa = 7.15WSDD342 pKa = 3.34EE343 pKa = 4.3GVVVASSGAFQYY355 pKa = 10.65FRR357 pKa = 11.84IGRR360 pKa = 11.84LFDD363 pKa = 3.18TTLYY367 pKa = 10.83YY368 pKa = 10.92AFPTSGTYY376 pKa = 10.66ARR378 pKa = 11.84DD379 pKa = 3.54DD380 pKa = 3.71PSALRR385 pKa = 11.84RR386 pKa = 11.84STSGDD391 pKa = 2.87LHH393 pKa = 6.92YY394 pKa = 10.88YY395 pKa = 9.82SPHH398 pKa = 5.8EE399 pKa = 4.1KK400 pKa = 10.41RR401 pKa = 11.84FVSYY405 pKa = 8.88TADD408 pKa = 3.26DD409 pKa = 3.35THH411 pKa = 7.43YY412 pKa = 10.49HH413 pKa = 5.12VFGVSVPRR421 pKa = 11.84SLADD425 pKa = 3.09YY426 pKa = 10.3CAATFCRR433 pKa = 11.84AVRR436 pKa = 11.84DD437 pKa = 3.89DD438 pKa = 4.25KK439 pKa = 11.41FYY441 pKa = 11.4DD442 pKa = 3.6SLRR445 pKa = 11.84SYY447 pKa = 9.17YY448 pKa = 9.02QNRR451 pKa = 11.84CRR453 pKa = 11.84AIGFSDD459 pKa = 4.19ARR461 pKa = 11.84DD462 pKa = 3.46TLMLDD467 pKa = 4.82FIIHH471 pKa = 6.59LCDD474 pKa = 3.9DD475 pKa = 3.77ASLRR479 pKa = 11.84TFGFSRR485 pKa = 11.84LSSAPSSWSAYY496 pKa = 7.55CLSWILVKK504 pKa = 10.52VNHH507 pKa = 5.93MMPLALTSYY516 pKa = 10.11VLNVLHH522 pKa = 7.06RR523 pKa = 11.84FFGAKK528 pKa = 8.97AAPWNWATIHH538 pKa = 6.56LPTYY542 pKa = 11.42DD543 pKa = 3.23MVTSPFRR550 pKa = 11.84LRR552 pKa = 11.84LFGRR556 pKa = 11.84NPTVFNLEE564 pKa = 3.89RR565 pKa = 11.84FRR567 pKa = 11.84AEE569 pKa = 3.62ATVAGSPDD577 pKa = 3.38RR578 pKa = 11.84GQSAEE583 pKa = 4.11GASQDD588 pKa = 3.07HH589 pKa = 6.86HH590 pKa = 7.01EE591 pKa = 4.42YY592 pKa = 10.56DD593 pKa = 4.24IKK595 pKa = 11.45SCDD598 pKa = 3.22ASVASRR604 pKa = 11.84SSSTPLPGDD613 pKa = 3.37SSTSGSVLLLDD624 pKa = 3.87DD625 pKa = 3.41TAGRR629 pKa = 11.84VHH631 pKa = 7.09EE632 pKa = 5.38DD633 pKa = 3.27DD634 pKa = 3.52SSARR638 pKa = 11.84STPHH642 pKa = 6.49RR643 pKa = 11.84GSGILSNKK651 pKa = 9.12SKK653 pKa = 10.8ARR655 pKa = 11.84RR656 pKa = 11.84KK657 pKa = 7.45SHH659 pKa = 6.72RR660 pKa = 11.84PSHH663 pKa = 6.84DD664 pKa = 3.33NSDD667 pKa = 3.59TDD669 pKa = 3.04HH670 pKa = 7.0GYY672 pKa = 10.32SVCHH676 pKa = 6.51PKK678 pKa = 9.4RR679 pKa = 11.84TTHH682 pKa = 6.68PLCPDD687 pKa = 3.63PTASCGPHH695 pKa = 6.48FFMSVCEE702 pKa = 4.39SNEE705 pKa = 4.09SVPTLFHH712 pKa = 5.9AHH714 pKa = 5.59SVGGEE719 pKa = 4.31DD720 pKa = 2.85ITHH723 pKa = 5.71VVDD726 pKa = 3.83PALGASISQRR736 pKa = 11.84FSASQLRR743 pKa = 11.84LLGWSIDD750 pKa = 4.11GIFNTLSGAATSSFVEE766 pKa = 4.32STLLCLSRR774 pKa = 11.84FMRR777 pKa = 11.84EE778 pKa = 4.01VPPTQPVTEE787 pKa = 4.23ARR789 pKa = 11.84RR790 pKa = 11.84SLLFCRR796 pKa = 11.84PRR798 pKa = 11.84EE799 pKa = 3.95RR800 pKa = 11.84FKK802 pKa = 11.25GYY804 pKa = 8.68DD805 pKa = 3.07TFDD808 pKa = 3.03IGFMGIAVPSSKK820 pKa = 9.77TKK822 pKa = 10.8GIEE825 pKa = 3.61ACLRR829 pKa = 11.84EE830 pKa = 4.46VARR833 pKa = 11.84QHH835 pKa = 6.53SRR837 pKa = 11.84ADD839 pKa = 3.7LPNEE843 pKa = 4.11GAA845 pKa = 4.15
MM1 pKa = 7.32KK2 pKa = 10.52VVFLVSFAVFLICQVPPVSYY22 pKa = 9.91WLEE25 pKa = 4.26DD26 pKa = 3.85LLDD29 pKa = 4.76EE30 pKa = 4.59YY31 pKa = 11.57NTMCSYY37 pKa = 10.91EE38 pKa = 4.02YY39 pKa = 10.97AAAEE43 pKa = 4.11AYY45 pKa = 10.86NNFVHH50 pKa = 6.58KK51 pKa = 10.34HH52 pKa = 4.47RR53 pKa = 11.84VAAYY57 pKa = 8.32AAGVRR62 pKa = 11.84ILRR65 pKa = 11.84YY66 pKa = 7.27RR67 pKa = 11.84TPWYY71 pKa = 10.01CFASRR76 pKa = 11.84SVPVVHH82 pKa = 6.76PFSWLMGQYY91 pKa = 10.43DD92 pKa = 3.72ATIDD96 pKa = 3.47RR97 pKa = 11.84VGTLMQRR104 pKa = 11.84LEE106 pKa = 4.02IANVSISDD114 pKa = 4.68GIRR117 pKa = 11.84TVADD121 pKa = 3.65TALKK125 pKa = 10.44AYY127 pKa = 9.85FYY129 pKa = 10.87YY130 pKa = 10.4QIIWFQLFSIIMLVGFFSAVICSSRR155 pKa = 11.84VRR157 pKa = 11.84VVRR160 pKa = 11.84IRR162 pKa = 11.84NRR164 pKa = 11.84VRR166 pKa = 11.84GYY168 pKa = 10.63SVTEE172 pKa = 3.71LRR174 pKa = 11.84SDD176 pKa = 3.94FEE178 pKa = 4.49EE179 pKa = 4.56SMSAILAPLSRR190 pKa = 11.84GHH192 pKa = 6.3SCLNFQRR199 pKa = 11.84RR200 pKa = 11.84VVEE203 pKa = 4.36SWAIDD208 pKa = 3.18QLLRR212 pKa = 11.84YY213 pKa = 7.7FRR215 pKa = 11.84CFRR218 pKa = 11.84SVASSQGRR226 pKa = 11.84WAEE229 pKa = 3.97VGHH232 pKa = 6.34RR233 pKa = 11.84LHH235 pKa = 6.79RR236 pKa = 11.84CSPVVLDD243 pKa = 3.81GAFVPEE249 pKa = 3.7YY250 pKa = 10.1DD251 pKa = 4.07QRR253 pKa = 11.84FSACRR258 pKa = 11.84RR259 pKa = 11.84HH260 pKa = 6.64PSVCPDD266 pKa = 3.32RR267 pKa = 11.84FDD269 pKa = 3.29IPAAILSHH277 pKa = 6.53VDD279 pKa = 3.35YY280 pKa = 11.57YY281 pKa = 8.69MTPDD285 pKa = 3.22QLAAAVTGPTFIVNHH300 pKa = 7.26DD301 pKa = 3.8YY302 pKa = 11.66SSIDD306 pKa = 3.43TLSVAEE312 pKa = 4.05VSLRR316 pKa = 11.84SAGGLVTASVRR327 pKa = 11.84DD328 pKa = 3.95GPTFGPHH335 pKa = 7.18PYY337 pKa = 9.92YY338 pKa = 10.24HH339 pKa = 7.15WSDD342 pKa = 3.34EE343 pKa = 4.3GVVVASSGAFQYY355 pKa = 10.65FRR357 pKa = 11.84IGRR360 pKa = 11.84LFDD363 pKa = 3.18TTLYY367 pKa = 10.83YY368 pKa = 10.92AFPTSGTYY376 pKa = 10.66ARR378 pKa = 11.84DD379 pKa = 3.54DD380 pKa = 3.71PSALRR385 pKa = 11.84RR386 pKa = 11.84STSGDD391 pKa = 2.87LHH393 pKa = 6.92YY394 pKa = 10.88YY395 pKa = 9.82SPHH398 pKa = 5.8EE399 pKa = 4.1KK400 pKa = 10.41RR401 pKa = 11.84FVSYY405 pKa = 8.88TADD408 pKa = 3.26DD409 pKa = 3.35THH411 pKa = 7.43YY412 pKa = 10.49HH413 pKa = 5.12VFGVSVPRR421 pKa = 11.84SLADD425 pKa = 3.09YY426 pKa = 10.3CAATFCRR433 pKa = 11.84AVRR436 pKa = 11.84DD437 pKa = 3.89DD438 pKa = 4.25KK439 pKa = 11.41FYY441 pKa = 11.4DD442 pKa = 3.6SLRR445 pKa = 11.84SYY447 pKa = 9.17YY448 pKa = 9.02QNRR451 pKa = 11.84CRR453 pKa = 11.84AIGFSDD459 pKa = 4.19ARR461 pKa = 11.84DD462 pKa = 3.46TLMLDD467 pKa = 4.82FIIHH471 pKa = 6.59LCDD474 pKa = 3.9DD475 pKa = 3.77ASLRR479 pKa = 11.84TFGFSRR485 pKa = 11.84LSSAPSSWSAYY496 pKa = 7.55CLSWILVKK504 pKa = 10.52VNHH507 pKa = 5.93MMPLALTSYY516 pKa = 10.11VLNVLHH522 pKa = 7.06RR523 pKa = 11.84FFGAKK528 pKa = 8.97AAPWNWATIHH538 pKa = 6.56LPTYY542 pKa = 11.42DD543 pKa = 3.23MVTSPFRR550 pKa = 11.84LRR552 pKa = 11.84LFGRR556 pKa = 11.84NPTVFNLEE564 pKa = 3.89RR565 pKa = 11.84FRR567 pKa = 11.84AEE569 pKa = 3.62ATVAGSPDD577 pKa = 3.38RR578 pKa = 11.84GQSAEE583 pKa = 4.11GASQDD588 pKa = 3.07HH589 pKa = 6.86HH590 pKa = 7.01EE591 pKa = 4.42YY592 pKa = 10.56DD593 pKa = 4.24IKK595 pKa = 11.45SCDD598 pKa = 3.22ASVASRR604 pKa = 11.84SSSTPLPGDD613 pKa = 3.37SSTSGSVLLLDD624 pKa = 3.87DD625 pKa = 3.41TAGRR629 pKa = 11.84VHH631 pKa = 7.09EE632 pKa = 5.38DD633 pKa = 3.27DD634 pKa = 3.52SSARR638 pKa = 11.84STPHH642 pKa = 6.49RR643 pKa = 11.84GSGILSNKK651 pKa = 9.12SKK653 pKa = 10.8ARR655 pKa = 11.84RR656 pKa = 11.84KK657 pKa = 7.45SHH659 pKa = 6.72RR660 pKa = 11.84PSHH663 pKa = 6.84DD664 pKa = 3.33NSDD667 pKa = 3.59TDD669 pKa = 3.04HH670 pKa = 7.0GYY672 pKa = 10.32SVCHH676 pKa = 6.51PKK678 pKa = 9.4RR679 pKa = 11.84TTHH682 pKa = 6.68PLCPDD687 pKa = 3.63PTASCGPHH695 pKa = 6.48FFMSVCEE702 pKa = 4.39SNEE705 pKa = 4.09SVPTLFHH712 pKa = 5.9AHH714 pKa = 5.59SVGGEE719 pKa = 4.31DD720 pKa = 2.85ITHH723 pKa = 5.71VVDD726 pKa = 3.83PALGASISQRR736 pKa = 11.84FSASQLRR743 pKa = 11.84LLGWSIDD750 pKa = 4.11GIFNTLSGAATSSFVEE766 pKa = 4.32STLLCLSRR774 pKa = 11.84FMRR777 pKa = 11.84EE778 pKa = 4.01VPPTQPVTEE787 pKa = 4.23ARR789 pKa = 11.84RR790 pKa = 11.84SLLFCRR796 pKa = 11.84PRR798 pKa = 11.84EE799 pKa = 3.95RR800 pKa = 11.84FKK802 pKa = 11.25GYY804 pKa = 8.68DD805 pKa = 3.07TFDD808 pKa = 3.03IGFMGIAVPSSKK820 pKa = 9.77TKK822 pKa = 10.8GIEE825 pKa = 3.61ACLRR829 pKa = 11.84EE830 pKa = 4.46VARR833 pKa = 11.84QHH835 pKa = 6.53SRR837 pKa = 11.84ADD839 pKa = 3.7LPNEE843 pKa = 4.11GAA845 pKa = 4.15
Molecular weight: 94.64 kDa
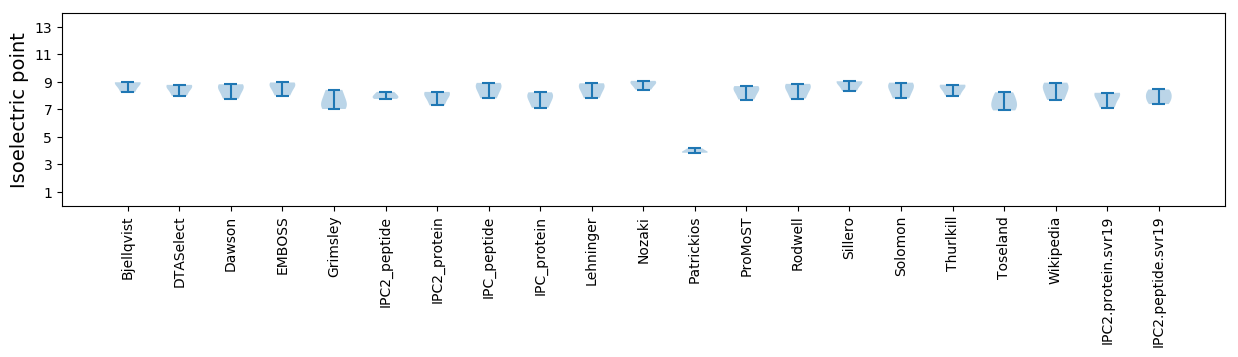
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A223DNF3|A0A223DNF3_9VIRU RNA-directed RNA polymerase OS=Lake Sinai virus 2 OX=1041831 PE=4 SV=1
MM1 pKa = 7.46NPPTTTTTTTRR12 pKa = 11.84TIRR15 pKa = 11.84APKK18 pKa = 9.53VQLTPNSATRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84NRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PAATIAGPLSIQPSSINRR55 pKa = 11.84RR56 pKa = 11.84MVSRR60 pKa = 11.84VTRR63 pKa = 11.84RR64 pKa = 11.84SAVISAAGLAWLRR77 pKa = 11.84QYY79 pKa = 10.78LNPMGPDD86 pKa = 3.32TTSVTGYY93 pKa = 9.4PDD95 pKa = 3.64GSAVTTCIADD105 pKa = 3.35YY106 pKa = 11.33SNTFNVSFPPRR117 pKa = 11.84EE118 pKa = 3.86ALYY121 pKa = 8.77CTGSSSSEE129 pKa = 3.99TPTLVDD135 pKa = 3.27ADD137 pKa = 4.14NYY139 pKa = 11.54AKK141 pKa = 9.73IDD143 pKa = 3.28KK144 pKa = 9.31WSSYY148 pKa = 11.15DD149 pKa = 3.01ITLCVLALPMLRR161 pKa = 11.84NVVMLRR167 pKa = 11.84LYY169 pKa = 9.99PHH171 pKa = 7.0TPTTFALTEE180 pKa = 3.89QTPNFPQRR188 pKa = 11.84FPNWSVYY195 pKa = 10.5SADD198 pKa = 3.01GTRR201 pKa = 11.84FNNGDD206 pKa = 3.53EE207 pKa = 4.19PGYY210 pKa = 9.36LQSYY214 pKa = 9.42VYY216 pKa = 10.53LPNVDD221 pKa = 3.44KK222 pKa = 10.99HH223 pKa = 6.81LSAARR228 pKa = 11.84GYY230 pKa = 10.17RR231 pKa = 11.84LLSRR235 pKa = 11.84GITGIFSAPALEE247 pKa = 4.3TQGFVTACQYY257 pKa = 10.68LAEE260 pKa = 4.9GSIQSQSIKK269 pKa = 10.29SDD271 pKa = 2.81AVRR274 pKa = 11.84SVTVNSDD281 pKa = 3.38GTVKK285 pKa = 10.52NVEE288 pKa = 4.34SSSQTVSSMPRR299 pKa = 11.84YY300 pKa = 9.32VFPLDD305 pKa = 4.14GDD307 pKa = 3.71NCAPSSLTEE316 pKa = 4.38TYY318 pKa = 9.68HH319 pKa = 6.29QAYY322 pKa = 8.98QSKK325 pKa = 8.3ATDD328 pKa = 3.65GFYY331 pKa = 10.3MPMLSSSRR339 pKa = 11.84DD340 pKa = 3.66NPFHH344 pKa = 6.81PPQPRR349 pKa = 11.84AIAVYY354 pKa = 10.4GSFLARR360 pKa = 11.84GCLDD364 pKa = 3.6PVSEE368 pKa = 4.19AHH370 pKa = 6.44EE371 pKa = 4.55ADD373 pKa = 4.37GPTHH377 pKa = 7.48DD378 pKa = 4.07IYY380 pKa = 11.27RR381 pKa = 11.84LNVADD386 pKa = 5.53DD387 pKa = 3.61VAPLFNTGVVWFEE400 pKa = 4.12GISPKK405 pKa = 10.28FSLKK409 pKa = 10.69LKK411 pKa = 9.41TRR413 pKa = 11.84TVLQYY418 pKa = 10.17IPTSGSVLANFTRR431 pKa = 11.84HH432 pKa = 5.55EE433 pKa = 4.0PTYY436 pKa = 11.03DD437 pKa = 4.04QIALDD442 pKa = 3.6AADD445 pKa = 4.17RR446 pKa = 11.84LRR448 pKa = 11.84NLMPHH453 pKa = 7.68AYY455 pKa = 8.52PAAYY459 pKa = 9.97NDD461 pKa = 3.89WGWLGDD467 pKa = 4.31LLDD470 pKa = 4.54SAISMLPGVGTVYY483 pKa = 10.97NIAKK487 pKa = 9.72PLIKK491 pKa = 9.96PAWNWLGNKK500 pKa = 9.42VSDD503 pKa = 3.96FFGNPVARR511 pKa = 11.84DD512 pKa = 3.01GDD514 pKa = 3.68IFFDD518 pKa = 3.96AKK520 pKa = 10.92
MM1 pKa = 7.46NPPTTTTTTTRR12 pKa = 11.84TIRR15 pKa = 11.84APKK18 pKa = 9.53VQLTPNSATRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84NRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84PAATIAGPLSIQPSSINRR55 pKa = 11.84RR56 pKa = 11.84MVSRR60 pKa = 11.84VTRR63 pKa = 11.84RR64 pKa = 11.84SAVISAAGLAWLRR77 pKa = 11.84QYY79 pKa = 10.78LNPMGPDD86 pKa = 3.32TTSVTGYY93 pKa = 9.4PDD95 pKa = 3.64GSAVTTCIADD105 pKa = 3.35YY106 pKa = 11.33SNTFNVSFPPRR117 pKa = 11.84EE118 pKa = 3.86ALYY121 pKa = 8.77CTGSSSSEE129 pKa = 3.99TPTLVDD135 pKa = 3.27ADD137 pKa = 4.14NYY139 pKa = 11.54AKK141 pKa = 9.73IDD143 pKa = 3.28KK144 pKa = 9.31WSSYY148 pKa = 11.15DD149 pKa = 3.01ITLCVLALPMLRR161 pKa = 11.84NVVMLRR167 pKa = 11.84LYY169 pKa = 9.99PHH171 pKa = 7.0TPTTFALTEE180 pKa = 3.89QTPNFPQRR188 pKa = 11.84FPNWSVYY195 pKa = 10.5SADD198 pKa = 3.01GTRR201 pKa = 11.84FNNGDD206 pKa = 3.53EE207 pKa = 4.19PGYY210 pKa = 9.36LQSYY214 pKa = 9.42VYY216 pKa = 10.53LPNVDD221 pKa = 3.44KK222 pKa = 10.99HH223 pKa = 6.81LSAARR228 pKa = 11.84GYY230 pKa = 10.17RR231 pKa = 11.84LLSRR235 pKa = 11.84GITGIFSAPALEE247 pKa = 4.3TQGFVTACQYY257 pKa = 10.68LAEE260 pKa = 4.9GSIQSQSIKK269 pKa = 10.29SDD271 pKa = 2.81AVRR274 pKa = 11.84SVTVNSDD281 pKa = 3.38GTVKK285 pKa = 10.52NVEE288 pKa = 4.34SSSQTVSSMPRR299 pKa = 11.84YY300 pKa = 9.32VFPLDD305 pKa = 4.14GDD307 pKa = 3.71NCAPSSLTEE316 pKa = 4.38TYY318 pKa = 9.68HH319 pKa = 6.29QAYY322 pKa = 8.98QSKK325 pKa = 8.3ATDD328 pKa = 3.65GFYY331 pKa = 10.3MPMLSSSRR339 pKa = 11.84DD340 pKa = 3.66NPFHH344 pKa = 6.81PPQPRR349 pKa = 11.84AIAVYY354 pKa = 10.4GSFLARR360 pKa = 11.84GCLDD364 pKa = 3.6PVSEE368 pKa = 4.19AHH370 pKa = 6.44EE371 pKa = 4.55ADD373 pKa = 4.37GPTHH377 pKa = 7.48DD378 pKa = 4.07IYY380 pKa = 11.27RR381 pKa = 11.84LNVADD386 pKa = 5.53DD387 pKa = 3.61VAPLFNTGVVWFEE400 pKa = 4.12GISPKK405 pKa = 10.28FSLKK409 pKa = 10.69LKK411 pKa = 9.41TRR413 pKa = 11.84TVLQYY418 pKa = 10.17IPTSGSVLANFTRR431 pKa = 11.84HH432 pKa = 5.55EE433 pKa = 4.0PTYY436 pKa = 11.03DD437 pKa = 4.04QIALDD442 pKa = 3.6AADD445 pKa = 4.17RR446 pKa = 11.84LRR448 pKa = 11.84NLMPHH453 pKa = 7.68AYY455 pKa = 8.52PAAYY459 pKa = 9.97NDD461 pKa = 3.89WGWLGDD467 pKa = 4.31LLDD470 pKa = 4.54SAISMLPGVGTVYY483 pKa = 10.97NIAKK487 pKa = 9.72PLIKK491 pKa = 9.96PAWNWLGNKK500 pKa = 9.42VSDD503 pKa = 3.96FFGNPVARR511 pKa = 11.84DD512 pKa = 3.01GDD514 pKa = 3.68IFFDD518 pKa = 3.96AKK520 pKa = 10.92
Molecular weight: 57.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1987 |
520 |
845 |
662.3 |
74.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.455 ± 0.288 | 2.416 ± 0.404 |
5.788 ± 0.332 | 3.221 ± 0.279 |
4.58 ± 0.622 | 5.033 ± 0.457 |
2.768 ± 0.565 | 4.479 ± 0.457 |
2.315 ± 0.271 | 9.26 ± 1.297 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.912 ± 0.152 | 2.969 ± 0.663 |
6.543 ± 0.667 | 2.567 ± 0.219 |
8.052 ± 0.34 | 10.216 ± 0.708 |
6.442 ± 0.674 | 6.794 ± 0.565 |
1.409 ± 0.051 | 4.781 ± 0.211 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
