
Gremmeniella abietina RNA virus MS2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Gammapartitivirus; Gremmeniella abietina RNA virus MS1
Average proteome isoelectric point is 8.0
Get precalculated fractions of proteins
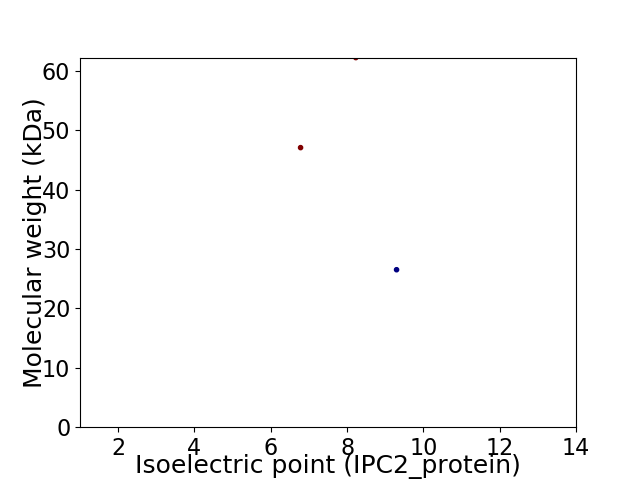
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6GYB9|Q6GYB9_9VIRU Putative RNA-dependent RNA polymerase OS=Gremmeniella abietina RNA virus MS2 OX=279540 PE=4 SV=1
MM1 pKa = 7.97SDD3 pKa = 3.28RR4 pKa = 11.84QSEE7 pKa = 4.39ISSSVAPGDD16 pKa = 4.09SVSSRR21 pKa = 11.84GGKK24 pKa = 9.03KK25 pKa = 10.03GKK27 pKa = 9.08PGKK30 pKa = 10.24AEE32 pKa = 3.6RR33 pKa = 11.84AARR36 pKa = 11.84RR37 pKa = 11.84AGQGSTPGVAASSAKK52 pKa = 10.49ASLFSSAGQADD63 pKa = 4.05INPTPGKK70 pKa = 9.98FPVVFATGAGEE81 pKa = 3.97PTRR84 pKa = 11.84DD85 pKa = 3.33AEE87 pKa = 4.15FSYY90 pKa = 11.18DD91 pKa = 3.32FSAVKK96 pKa = 10.17RR97 pKa = 11.84CVVGFGDD104 pKa = 4.52RR105 pKa = 11.84YY106 pKa = 10.25VKK108 pKa = 10.19NARR111 pKa = 11.84YY112 pKa = 9.93SEE114 pKa = 4.46FKK116 pKa = 10.84ANSGVADD123 pKa = 3.91SAFKK127 pKa = 10.68CYY129 pKa = 10.0LAASCLLGLAQQTVHH144 pKa = 6.1AHH146 pKa = 4.87VNMGLPQGDD155 pKa = 4.21FAPVSSSEE163 pKa = 4.01VLNFTAVKK171 pKa = 10.26AYY173 pKa = 9.48IGQFGEE179 pKa = 4.31FAVPSTGTRR188 pKa = 11.84YY189 pKa = 8.97MLADD193 pKa = 3.77YY194 pKa = 10.85QSTVCKK200 pKa = 10.17LVYY203 pKa = 10.39LADD206 pKa = 3.61KK207 pKa = 10.53CLRR210 pKa = 11.84DD211 pKa = 3.77ANHH214 pKa = 5.77NTVLSRR220 pKa = 11.84AWLPMSSGDD229 pKa = 3.22RR230 pKa = 11.84TTKK233 pKa = 10.5VVIAAALSQLLSNAEE248 pKa = 4.15LKK250 pKa = 10.56ISSTVLEE257 pKa = 4.49DD258 pKa = 3.48AVLSGDD264 pKa = 5.79VPDD267 pKa = 3.92AWNAVKK273 pKa = 9.38TVFGDD278 pKa = 3.61APGEE282 pKa = 4.22GVPDD286 pKa = 3.31ARR288 pKa = 11.84DD289 pKa = 3.22RR290 pKa = 11.84FDD292 pKa = 5.59FLFKK296 pKa = 10.7SYY298 pKa = 11.52ADD300 pKa = 3.29VGQFTTAFTTGAASAVLAEE319 pKa = 4.72LHH321 pKa = 6.7LPWSSPSAGHH331 pKa = 7.38LDD333 pKa = 3.04WDD335 pKa = 4.46YY336 pKa = 11.79NPKK339 pKa = 9.1TRR341 pKa = 11.84FSALADD347 pKa = 2.99SWARR351 pKa = 11.84ISASYY356 pKa = 10.96SGFFHH361 pKa = 6.48MASGLSDD368 pKa = 3.5RR369 pKa = 11.84SNALGSEE376 pKa = 4.29SQFVEE381 pKa = 4.12VRR383 pKa = 11.84SIDD386 pKa = 3.46SVTVVKK392 pKa = 9.9TFLALSAPQFSLAACFPPEE411 pKa = 4.4CIFVGSLDD419 pKa = 3.39RR420 pKa = 11.84RR421 pKa = 11.84VVVTTPIPVSQRR433 pKa = 11.84ATEE436 pKa = 4.0FCLRR440 pKa = 11.84DD441 pKa = 3.44WKK443 pKa = 11.18
MM1 pKa = 7.97SDD3 pKa = 3.28RR4 pKa = 11.84QSEE7 pKa = 4.39ISSSVAPGDD16 pKa = 4.09SVSSRR21 pKa = 11.84GGKK24 pKa = 9.03KK25 pKa = 10.03GKK27 pKa = 9.08PGKK30 pKa = 10.24AEE32 pKa = 3.6RR33 pKa = 11.84AARR36 pKa = 11.84RR37 pKa = 11.84AGQGSTPGVAASSAKK52 pKa = 10.49ASLFSSAGQADD63 pKa = 4.05INPTPGKK70 pKa = 9.98FPVVFATGAGEE81 pKa = 3.97PTRR84 pKa = 11.84DD85 pKa = 3.33AEE87 pKa = 4.15FSYY90 pKa = 11.18DD91 pKa = 3.32FSAVKK96 pKa = 10.17RR97 pKa = 11.84CVVGFGDD104 pKa = 4.52RR105 pKa = 11.84YY106 pKa = 10.25VKK108 pKa = 10.19NARR111 pKa = 11.84YY112 pKa = 9.93SEE114 pKa = 4.46FKK116 pKa = 10.84ANSGVADD123 pKa = 3.91SAFKK127 pKa = 10.68CYY129 pKa = 10.0LAASCLLGLAQQTVHH144 pKa = 6.1AHH146 pKa = 4.87VNMGLPQGDD155 pKa = 4.21FAPVSSSEE163 pKa = 4.01VLNFTAVKK171 pKa = 10.26AYY173 pKa = 9.48IGQFGEE179 pKa = 4.31FAVPSTGTRR188 pKa = 11.84YY189 pKa = 8.97MLADD193 pKa = 3.77YY194 pKa = 10.85QSTVCKK200 pKa = 10.17LVYY203 pKa = 10.39LADD206 pKa = 3.61KK207 pKa = 10.53CLRR210 pKa = 11.84DD211 pKa = 3.77ANHH214 pKa = 5.77NTVLSRR220 pKa = 11.84AWLPMSSGDD229 pKa = 3.22RR230 pKa = 11.84TTKK233 pKa = 10.5VVIAAALSQLLSNAEE248 pKa = 4.15LKK250 pKa = 10.56ISSTVLEE257 pKa = 4.49DD258 pKa = 3.48AVLSGDD264 pKa = 5.79VPDD267 pKa = 3.92AWNAVKK273 pKa = 9.38TVFGDD278 pKa = 3.61APGEE282 pKa = 4.22GVPDD286 pKa = 3.31ARR288 pKa = 11.84DD289 pKa = 3.22RR290 pKa = 11.84FDD292 pKa = 5.59FLFKK296 pKa = 10.7SYY298 pKa = 11.52ADD300 pKa = 3.29VGQFTTAFTTGAASAVLAEE319 pKa = 4.72LHH321 pKa = 6.7LPWSSPSAGHH331 pKa = 7.38LDD333 pKa = 3.04WDD335 pKa = 4.46YY336 pKa = 11.79NPKK339 pKa = 9.1TRR341 pKa = 11.84FSALADD347 pKa = 2.99SWARR351 pKa = 11.84ISASYY356 pKa = 10.96SGFFHH361 pKa = 6.48MASGLSDD368 pKa = 3.5RR369 pKa = 11.84SNALGSEE376 pKa = 4.29SQFVEE381 pKa = 4.12VRR383 pKa = 11.84SIDD386 pKa = 3.46SVTVVKK392 pKa = 9.9TFLALSAPQFSLAACFPPEE411 pKa = 4.4CIFVGSLDD419 pKa = 3.39RR420 pKa = 11.84RR421 pKa = 11.84VVVTTPIPVSQRR433 pKa = 11.84ATEE436 pKa = 4.0FCLRR440 pKa = 11.84DD441 pKa = 3.44WKK443 pKa = 11.18
Molecular weight: 47.1 kDa
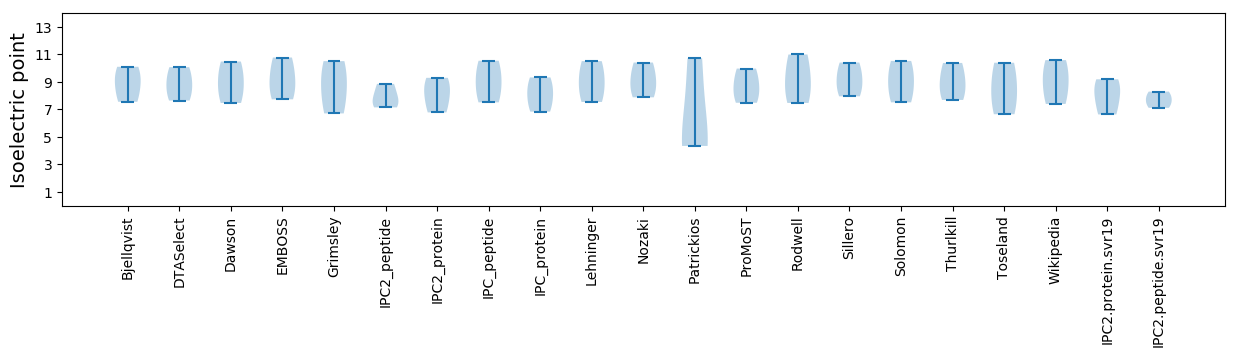
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6GYB8|Q6GYB8_9VIRU Coat protein OS=Gremmeniella abietina RNA virus MS2 OX=279540 PE=4 SV=1
MM1 pKa = 7.54SKK3 pKa = 7.84THH5 pKa = 6.11TNLVGVKK12 pKa = 10.16SSPPKK17 pKa = 8.94PKK19 pKa = 10.81GNIRR23 pKa = 11.84QPSASQLRR31 pKa = 11.84FKK33 pKa = 10.66KK34 pKa = 10.46ALARR38 pKa = 11.84ALSVRR43 pKa = 11.84SKK45 pKa = 10.19FLKK48 pKa = 10.18RR49 pKa = 11.84RR50 pKa = 11.84LSKK53 pKa = 11.01PEE55 pKa = 3.66FVQLSAAVKK64 pKa = 9.93KK65 pKa = 10.32QIKK68 pKa = 9.93QEE70 pKa = 4.05TTPVNRR76 pKa = 11.84RR77 pKa = 11.84LNRR80 pKa = 11.84VIDD83 pKa = 3.31IVVRR87 pKa = 11.84LEE89 pKa = 3.65EE90 pKa = 4.29AANSSLSCLDD100 pKa = 3.8RR101 pKa = 11.84PALDD105 pKa = 3.25AAVRR109 pKa = 11.84NLYY112 pKa = 10.43DD113 pKa = 3.28RR114 pKa = 11.84LNLVRR119 pKa = 11.84SNLLEE124 pKa = 3.96AMRR127 pKa = 11.84EE128 pKa = 4.1SSSLSEE134 pKa = 4.42GCSHH138 pKa = 6.67VSKK141 pKa = 10.68KK142 pKa = 10.58SVIHH146 pKa = 6.03LPEE149 pKa = 4.08PVKK152 pKa = 10.62VDD154 pKa = 3.57KK155 pKa = 10.8IFPGFGCISARR166 pKa = 11.84IPSGRR171 pKa = 11.84QVLCCSRR178 pKa = 11.84CSGLRR183 pKa = 11.84NDD185 pKa = 6.09AFFEE189 pKa = 4.3WSGLQPSFIDD199 pKa = 3.8KK200 pKa = 9.76EE201 pKa = 4.27GHH203 pKa = 5.97SMVEE207 pKa = 4.49AYY209 pKa = 8.39PLPKK213 pKa = 9.9SIKK216 pKa = 10.09NISPLDD222 pKa = 3.82FQHH225 pKa = 5.9VWTSVLDD232 pKa = 3.16IHH234 pKa = 7.22YY235 pKa = 10.91SRR237 pKa = 5.53
MM1 pKa = 7.54SKK3 pKa = 7.84THH5 pKa = 6.11TNLVGVKK12 pKa = 10.16SSPPKK17 pKa = 8.94PKK19 pKa = 10.81GNIRR23 pKa = 11.84QPSASQLRR31 pKa = 11.84FKK33 pKa = 10.66KK34 pKa = 10.46ALARR38 pKa = 11.84ALSVRR43 pKa = 11.84SKK45 pKa = 10.19FLKK48 pKa = 10.18RR49 pKa = 11.84RR50 pKa = 11.84LSKK53 pKa = 11.01PEE55 pKa = 3.66FVQLSAAVKK64 pKa = 9.93KK65 pKa = 10.32QIKK68 pKa = 9.93QEE70 pKa = 4.05TTPVNRR76 pKa = 11.84RR77 pKa = 11.84LNRR80 pKa = 11.84VIDD83 pKa = 3.31IVVRR87 pKa = 11.84LEE89 pKa = 3.65EE90 pKa = 4.29AANSSLSCLDD100 pKa = 3.8RR101 pKa = 11.84PALDD105 pKa = 3.25AAVRR109 pKa = 11.84NLYY112 pKa = 10.43DD113 pKa = 3.28RR114 pKa = 11.84LNLVRR119 pKa = 11.84SNLLEE124 pKa = 3.96AMRR127 pKa = 11.84EE128 pKa = 4.1SSSLSEE134 pKa = 4.42GCSHH138 pKa = 6.67VSKK141 pKa = 10.68KK142 pKa = 10.58SVIHH146 pKa = 6.03LPEE149 pKa = 4.08PVKK152 pKa = 10.62VDD154 pKa = 3.57KK155 pKa = 10.8IFPGFGCISARR166 pKa = 11.84IPSGRR171 pKa = 11.84QVLCCSRR178 pKa = 11.84CSGLRR183 pKa = 11.84NDD185 pKa = 6.09AFFEE189 pKa = 4.3WSGLQPSFIDD199 pKa = 3.8KK200 pKa = 9.76EE201 pKa = 4.27GHH203 pKa = 5.97SMVEE207 pKa = 4.49AYY209 pKa = 8.39PLPKK213 pKa = 9.9SIKK216 pKa = 10.09NISPLDD222 pKa = 3.82FQHH225 pKa = 5.9VWTSVLDD232 pKa = 3.16IHH234 pKa = 7.22YY235 pKa = 10.91SRR237 pKa = 5.53
Molecular weight: 26.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1219 |
237 |
539 |
406.3 |
45.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.368 ± 2.195 | 1.723 ± 0.324 |
5.414 ± 0.559 | 5.086 ± 0.947 |
5.168 ± 0.533 | 7.055 ± 1.155 |
1.559 ± 0.336 | 3.035 ± 0.718 |
5.742 ± 0.82 | 7.875 ± 0.917 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.133 ± 0.712 | 3.035 ± 0.415 |
5.25 ± 0.369 | 3.363 ± 0.216 |
7.137 ± 0.901 | 10.5 ± 1.541 |
4.594 ± 0.848 | 7.547 ± 0.731 |
2.297 ± 0.864 | 3.117 ± 0.881 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
