
Scytalidium lignicola (Hyphomycete)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Leotiomycetes;
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
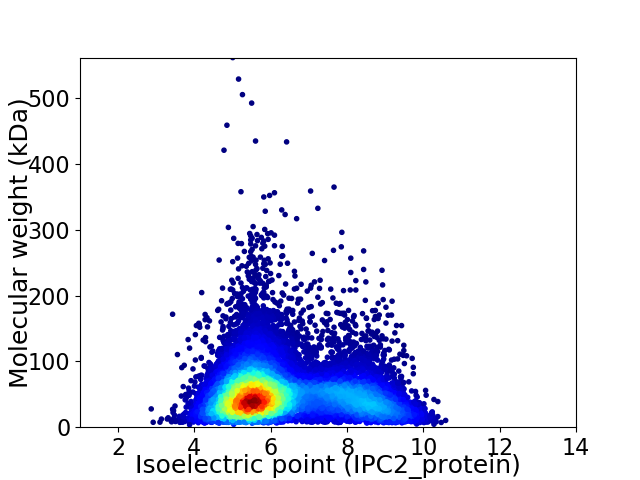
Virtual 2D-PAGE plot for 12794 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3E2HPT5|A0A3E2HPT5_SCYLI CID domain-containing protein (Fragment) OS=Scytalidium lignicola OX=5539 GN=B7463_g980 PE=4 SV=1
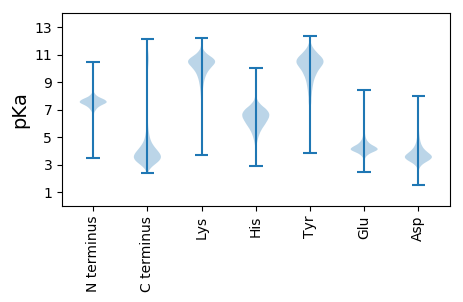
MM1 pKa = 7.33NCDD4 pKa = 3.5DD5 pKa = 4.43TRR7 pKa = 11.84TEE9 pKa = 4.43AIKK12 pKa = 9.83MALVKK17 pKa = 9.92VQNVAYY23 pKa = 10.06RR24 pKa = 11.84AYY26 pKa = 10.75DD27 pKa = 3.51RR28 pKa = 11.84LDD30 pKa = 3.52TLHH33 pKa = 7.45DD34 pKa = 4.47LLNVAQVQLSNFDD47 pKa = 3.67YY48 pKa = 11.37LEE50 pKa = 3.97WSTFNTFEE58 pKa = 4.05TFYY61 pKa = 11.23GQMLVDD67 pKa = 3.89TTRR70 pKa = 11.84TNGGVITEE78 pKa = 4.26TLNYY82 pKa = 9.81LDD84 pKa = 5.37EE85 pKa = 5.43IRR87 pKa = 11.84DD88 pKa = 3.67FLGNIVLAAQNVNADD103 pKa = 3.71QYY105 pKa = 11.57HH106 pKa = 6.5DD107 pKa = 4.13TLSLDD112 pKa = 3.53IWCDD116 pKa = 3.79SAWEE120 pKa = 4.26TTTDD124 pKa = 3.31NNGRR128 pKa = 11.84YY129 pKa = 9.15YY130 pKa = 10.8DD131 pKa = 4.0GPTDD135 pKa = 4.36VMVYY139 pKa = 9.53WDD141 pKa = 3.73GSGNNMGWIDD151 pKa = 4.1PPEE154 pKa = 4.72DD155 pKa = 3.29ATTPADD161 pKa = 6.1LILCTNSPLDD171 pKa = 3.76LGGYY175 pKa = 7.2TINHH179 pKa = 5.98DD180 pKa = 4.03TEE182 pKa = 4.45PVSVITICGDD192 pKa = 3.4ALDD195 pKa = 5.37DD196 pKa = 4.44MISNGMTLTSLGAGDD211 pKa = 4.38IKK213 pKa = 11.14DD214 pKa = 3.78GTDD217 pKa = 3.59LSDD220 pKa = 3.81EE221 pKa = 4.34VEE223 pKa = 4.74KK224 pKa = 10.91IPSFLLLHH232 pKa = 6.23EE233 pKa = 5.24LSHH236 pKa = 6.43AVSVMGRR243 pKa = 11.84GSNTIDD249 pKa = 3.65VKK251 pKa = 10.8FQNPTEE257 pKa = 4.12EE258 pKa = 4.57GVLNAYY264 pKa = 9.78GLEE267 pKa = 4.01NCILLAVNRR276 pKa = 11.84PDD278 pKa = 3.78LTIEE282 pKa = 4.16NADD285 pKa = 3.84TLALLAAALYY295 pKa = 9.05LQQKK299 pKa = 5.91TWWTGEE305 pKa = 4.06AEE307 pKa = 4.16DD308 pKa = 4.04LSYY311 pKa = 10.77RR312 pKa = 11.84ASPSIQFLPPGAAAPP327 pKa = 3.91
MM1 pKa = 7.33NCDD4 pKa = 3.5DD5 pKa = 4.43TRR7 pKa = 11.84TEE9 pKa = 4.43AIKK12 pKa = 9.83MALVKK17 pKa = 9.92VQNVAYY23 pKa = 10.06RR24 pKa = 11.84AYY26 pKa = 10.75DD27 pKa = 3.51RR28 pKa = 11.84LDD30 pKa = 3.52TLHH33 pKa = 7.45DD34 pKa = 4.47LLNVAQVQLSNFDD47 pKa = 3.67YY48 pKa = 11.37LEE50 pKa = 3.97WSTFNTFEE58 pKa = 4.05TFYY61 pKa = 11.23GQMLVDD67 pKa = 3.89TTRR70 pKa = 11.84TNGGVITEE78 pKa = 4.26TLNYY82 pKa = 9.81LDD84 pKa = 5.37EE85 pKa = 5.43IRR87 pKa = 11.84DD88 pKa = 3.67FLGNIVLAAQNVNADD103 pKa = 3.71QYY105 pKa = 11.57HH106 pKa = 6.5DD107 pKa = 4.13TLSLDD112 pKa = 3.53IWCDD116 pKa = 3.79SAWEE120 pKa = 4.26TTTDD124 pKa = 3.31NNGRR128 pKa = 11.84YY129 pKa = 9.15YY130 pKa = 10.8DD131 pKa = 4.0GPTDD135 pKa = 4.36VMVYY139 pKa = 9.53WDD141 pKa = 3.73GSGNNMGWIDD151 pKa = 4.1PPEE154 pKa = 4.72DD155 pKa = 3.29ATTPADD161 pKa = 6.1LILCTNSPLDD171 pKa = 3.76LGGYY175 pKa = 7.2TINHH179 pKa = 5.98DD180 pKa = 4.03TEE182 pKa = 4.45PVSVITICGDD192 pKa = 3.4ALDD195 pKa = 5.37DD196 pKa = 4.44MISNGMTLTSLGAGDD211 pKa = 4.38IKK213 pKa = 11.14DD214 pKa = 3.78GTDD217 pKa = 3.59LSDD220 pKa = 3.81EE221 pKa = 4.34VEE223 pKa = 4.74KK224 pKa = 10.91IPSFLLLHH232 pKa = 6.23EE233 pKa = 5.24LSHH236 pKa = 6.43AVSVMGRR243 pKa = 11.84GSNTIDD249 pKa = 3.65VKK251 pKa = 10.8FQNPTEE257 pKa = 4.12EE258 pKa = 4.57GVLNAYY264 pKa = 9.78GLEE267 pKa = 4.01NCILLAVNRR276 pKa = 11.84PDD278 pKa = 3.78LTIEE282 pKa = 4.16NADD285 pKa = 3.84TLALLAAALYY295 pKa = 9.05LQQKK299 pKa = 5.91TWWTGEE305 pKa = 4.06AEE307 pKa = 4.16DD308 pKa = 4.04LSYY311 pKa = 10.77RR312 pKa = 11.84ASPSIQFLPPGAAAPP327 pKa = 3.91
Molecular weight: 36.03 kDa
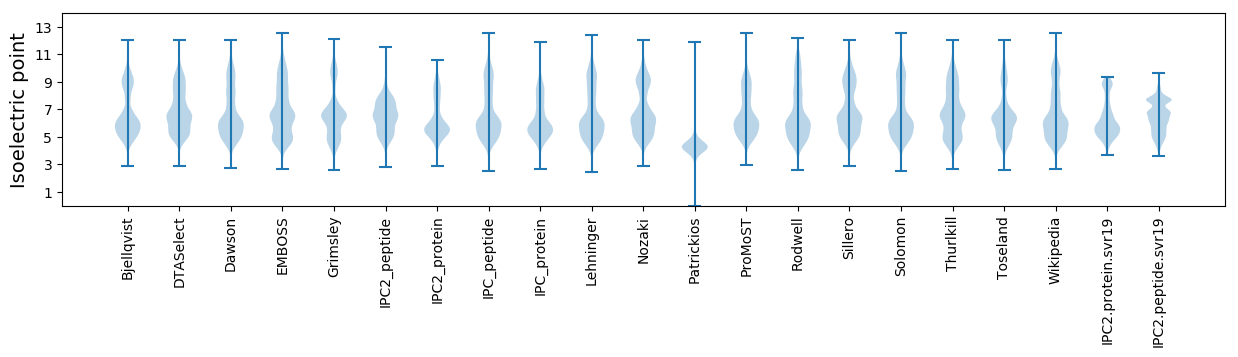
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3E2HHG2|A0A3E2HHG2_SCYLI Uncharacterized protein (Fragment) OS=Scytalidium lignicola OX=5539 GN=B7463_g3444 PE=3 SV=1
MM1 pKa = 7.73ILIRR5 pKa = 11.84IVQPRR10 pKa = 11.84LEE12 pKa = 4.36EE13 pKa = 3.86QDD15 pKa = 3.33QLLMQRR21 pKa = 11.84RR22 pKa = 11.84SEE24 pKa = 3.98RR25 pKa = 11.84DD26 pKa = 2.89RR27 pKa = 11.84KK28 pKa = 10.66NGLRR32 pKa = 11.84QKK34 pKa = 9.86SVRR37 pKa = 11.84YY38 pKa = 9.93DD39 pKa = 2.99IGTVQYY45 pKa = 10.16FSVANLPTTSGQAEE59 pKa = 4.48LTTSEE64 pKa = 4.2TLKK67 pKa = 10.48QLRR70 pKa = 11.84LKK72 pKa = 10.97ALGTGISGQSSDD84 pKa = 3.48YY85 pKa = 10.46TVISRR90 pKa = 11.84AIGHH94 pKa = 6.46RR95 pKa = 11.84PRR97 pKa = 11.84NGLGFGGGDD106 pKa = 4.46DD107 pKa = 5.22DD108 pKa = 6.84DD109 pKa = 5.51NNQTRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84ASGLFCFVGNWGFVCKK132 pKa = 10.31QRR134 pKa = 11.84DD135 pKa = 3.61GMDD138 pKa = 3.6CGLVFLPQSDD148 pKa = 3.79DD149 pKa = 4.47LAWQPAPKK157 pKa = 9.53RR158 pKa = 11.84RR159 pKa = 11.84KK160 pKa = 9.01VVNFPSASLPPQLLSRR176 pKa = 11.84RR177 pKa = 11.84LRR179 pKa = 11.84RR180 pKa = 11.84PWTAAPASCVGG191 pKa = 3.39
MM1 pKa = 7.73ILIRR5 pKa = 11.84IVQPRR10 pKa = 11.84LEE12 pKa = 4.36EE13 pKa = 3.86QDD15 pKa = 3.33QLLMQRR21 pKa = 11.84RR22 pKa = 11.84SEE24 pKa = 3.98RR25 pKa = 11.84DD26 pKa = 2.89RR27 pKa = 11.84KK28 pKa = 10.66NGLRR32 pKa = 11.84QKK34 pKa = 9.86SVRR37 pKa = 11.84YY38 pKa = 9.93DD39 pKa = 2.99IGTVQYY45 pKa = 10.16FSVANLPTTSGQAEE59 pKa = 4.48LTTSEE64 pKa = 4.2TLKK67 pKa = 10.48QLRR70 pKa = 11.84LKK72 pKa = 10.97ALGTGISGQSSDD84 pKa = 3.48YY85 pKa = 10.46TVISRR90 pKa = 11.84AIGHH94 pKa = 6.46RR95 pKa = 11.84PRR97 pKa = 11.84NGLGFGGGDD106 pKa = 4.46DD107 pKa = 5.22DD108 pKa = 6.84DD109 pKa = 5.51NNQTRR114 pKa = 11.84RR115 pKa = 11.84RR116 pKa = 11.84ASGLFCFVGNWGFVCKK132 pKa = 10.31QRR134 pKa = 11.84DD135 pKa = 3.61GMDD138 pKa = 3.6CGLVFLPQSDD148 pKa = 3.79DD149 pKa = 4.47LAWQPAPKK157 pKa = 9.53RR158 pKa = 11.84RR159 pKa = 11.84KK160 pKa = 9.01VVNFPSASLPPQLLSRR176 pKa = 11.84RR177 pKa = 11.84LRR179 pKa = 11.84RR180 pKa = 11.84PWTAAPASCVGG191 pKa = 3.39
Molecular weight: 21.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6524524 |
37 |
4968 |
510.0 |
56.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.981 ± 0.017 | 1.227 ± 0.008 |
5.519 ± 0.015 | 6.25 ± 0.021 |
3.826 ± 0.012 | 6.758 ± 0.02 |
2.283 ± 0.008 | 5.666 ± 0.014 |
5.082 ± 0.021 | 9.029 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.129 ± 0.007 | 4.048 ± 0.011 |
5.709 ± 0.02 | 3.957 ± 0.014 |
5.613 ± 0.016 | 8.365 ± 0.023 |
6.106 ± 0.019 | 6.037 ± 0.014 |
1.478 ± 0.008 | 2.926 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
