
Pseudoroseicyclus aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Pseudoroseicyclus
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
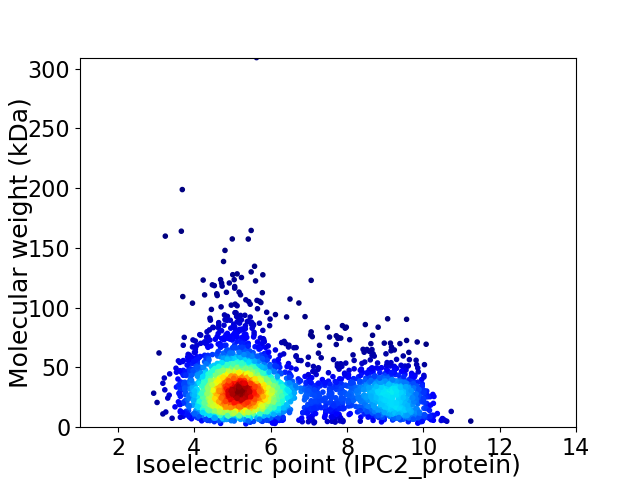
Virtual 2D-PAGE plot for 3295 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
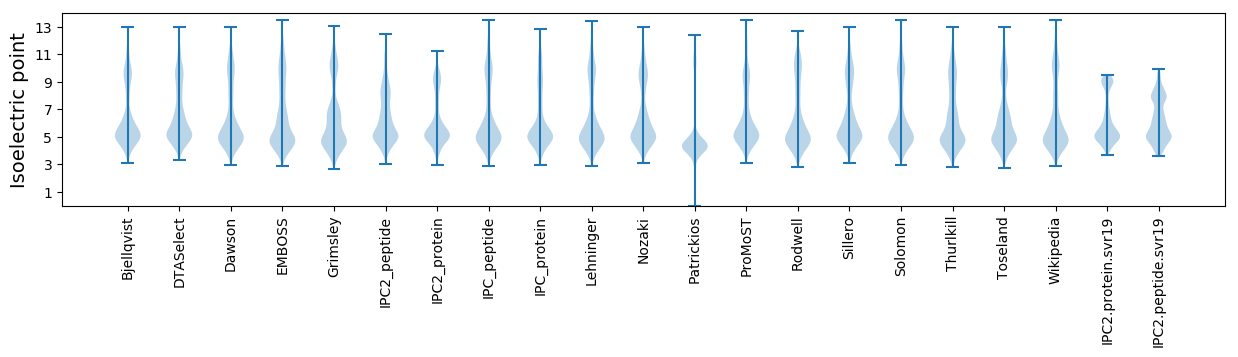
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318SVW2|A0A318SVW2_9RHOB YVTN family beta-propeller protein OS=Pseudoroseicyclus aestuarii OX=1795041 GN=DFP88_102286 PE=4 SV=1
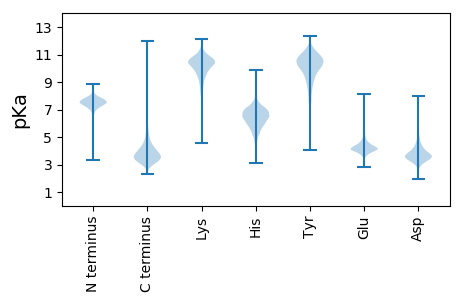
MM1 pKa = 7.07MRR3 pKa = 11.84TSIALLAALGLGSAATAQDD22 pKa = 3.75LSIATGGTGGTYY34 pKa = 9.96YY35 pKa = 10.36PYY37 pKa = 11.05GGALAEE43 pKa = 4.74LIGAHH48 pKa = 5.99VDD50 pKa = 3.39GASAVAEE57 pKa = 4.11VTGASVEE64 pKa = 3.99NMALIAQEE72 pKa = 3.83QSDD75 pKa = 4.06IAFVLADD82 pKa = 3.52TAYY85 pKa = 10.12GAVNGTIDD93 pKa = 4.05PFTDD97 pKa = 3.16RR98 pKa = 11.84AVDD101 pKa = 4.02GQALASIYY109 pKa = 9.94PNAVHH114 pKa = 7.09IVTLAGSGIEE124 pKa = 4.04SLQDD128 pKa = 3.43LVGKK132 pKa = 9.61RR133 pKa = 11.84VSVGAPGSGTEE144 pKa = 4.11VNAEE148 pKa = 3.94QLLSANGITYY158 pKa = 10.73DD159 pKa = 5.48DD160 pKa = 4.17IDD162 pKa = 3.8EE163 pKa = 4.09QRR165 pKa = 11.84LNFNEE170 pKa = 4.05TADD173 pKa = 3.72ALRR176 pKa = 11.84DD177 pKa = 3.49GDD179 pKa = 3.62IDD181 pKa = 3.85AGIWSVGPPTSSILNLAATTDD202 pKa = 3.39IAFVPLTEE210 pKa = 4.44EE211 pKa = 3.93EE212 pKa = 4.17VAAAQEE218 pKa = 4.17VQPVFAAYY226 pKa = 9.15EE227 pKa = 4.09LRR229 pKa = 11.84AGTYY233 pKa = 10.2DD234 pKa = 3.61GVDD237 pKa = 3.14EE238 pKa = 4.75GVATIAIPNVLVVNAAMDD256 pKa = 3.73EE257 pKa = 4.07EE258 pKa = 4.5LAYY261 pKa = 10.7QITSAIFEE269 pKa = 4.56HH270 pKa = 6.63TDD272 pKa = 3.03EE273 pKa = 5.69LIAVHH278 pKa = 6.76PAAEE282 pKa = 4.21DD283 pKa = 3.64TTVDD287 pKa = 3.94FSVNSTPIPFHH298 pKa = 6.81PGAIRR303 pKa = 11.84YY304 pKa = 8.34YY305 pKa = 10.34DD306 pKa = 3.49EE307 pKa = 4.57VGAEE311 pKa = 4.04VSDD314 pKa = 3.78AQRR317 pKa = 11.84PP318 pKa = 3.58
MM1 pKa = 7.07MRR3 pKa = 11.84TSIALLAALGLGSAATAQDD22 pKa = 3.75LSIATGGTGGTYY34 pKa = 9.96YY35 pKa = 10.36PYY37 pKa = 11.05GGALAEE43 pKa = 4.74LIGAHH48 pKa = 5.99VDD50 pKa = 3.39GASAVAEE57 pKa = 4.11VTGASVEE64 pKa = 3.99NMALIAQEE72 pKa = 3.83QSDD75 pKa = 4.06IAFVLADD82 pKa = 3.52TAYY85 pKa = 10.12GAVNGTIDD93 pKa = 4.05PFTDD97 pKa = 3.16RR98 pKa = 11.84AVDD101 pKa = 4.02GQALASIYY109 pKa = 9.94PNAVHH114 pKa = 7.09IVTLAGSGIEE124 pKa = 4.04SLQDD128 pKa = 3.43LVGKK132 pKa = 9.61RR133 pKa = 11.84VSVGAPGSGTEE144 pKa = 4.11VNAEE148 pKa = 3.94QLLSANGITYY158 pKa = 10.73DD159 pKa = 5.48DD160 pKa = 4.17IDD162 pKa = 3.8EE163 pKa = 4.09QRR165 pKa = 11.84LNFNEE170 pKa = 4.05TADD173 pKa = 3.72ALRR176 pKa = 11.84DD177 pKa = 3.49GDD179 pKa = 3.62IDD181 pKa = 3.85AGIWSVGPPTSSILNLAATTDD202 pKa = 3.39IAFVPLTEE210 pKa = 4.44EE211 pKa = 3.93EE212 pKa = 4.17VAAAQEE218 pKa = 4.17VQPVFAAYY226 pKa = 9.15EE227 pKa = 4.09LRR229 pKa = 11.84AGTYY233 pKa = 10.2DD234 pKa = 3.61GVDD237 pKa = 3.14EE238 pKa = 4.75GVATIAIPNVLVVNAAMDD256 pKa = 3.73EE257 pKa = 4.07EE258 pKa = 4.5LAYY261 pKa = 10.7QITSAIFEE269 pKa = 4.56HH270 pKa = 6.63TDD272 pKa = 3.03EE273 pKa = 5.69LIAVHH278 pKa = 6.76PAAEE282 pKa = 4.21DD283 pKa = 3.64TTVDD287 pKa = 3.94FSVNSTPIPFHH298 pKa = 6.81PGAIRR303 pKa = 11.84YY304 pKa = 8.34YY305 pKa = 10.34DD306 pKa = 3.49EE307 pKa = 4.57VGAEE311 pKa = 4.04VSDD314 pKa = 3.78AQRR317 pKa = 11.84PP318 pKa = 3.58
Molecular weight: 32.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318SQB0|A0A318SQB0_9RHOB Threonine/homoserine/homoserine lactone efflux protein OS=Pseudoroseicyclus aestuarii OX=1795041 GN=DFP88_104295 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97VLSAA44 pKa = 4.11
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1035711 |
26 |
2801 |
314.3 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.664 ± 0.079 | 0.781 ± 0.014 |
5.77 ± 0.042 | 6.332 ± 0.045 |
3.289 ± 0.03 | 9.311 ± 0.052 |
1.877 ± 0.023 | 4.551 ± 0.032 |
2.116 ± 0.038 | 10.68 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.598 ± 0.021 | 1.972 ± 0.025 |
5.599 ± 0.039 | 3.304 ± 0.025 |
7.507 ± 0.048 | 4.846 ± 0.032 |
5.269 ± 0.031 | 7.037 ± 0.033 |
1.452 ± 0.02 | 2.045 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
