
Cryobacterium sp. Hh4
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium; unclassified Cryobacterium
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
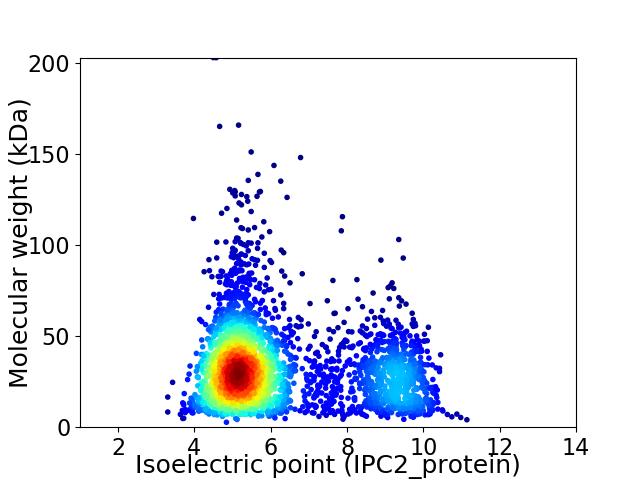
Virtual 2D-PAGE plot for 3130 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R9AZC9|A0A4R9AZC9_9MICO VOC family protein OS=Cryobacterium sp. Hh4 OX=1259157 GN=E3T48_14795 PE=4 SV=1
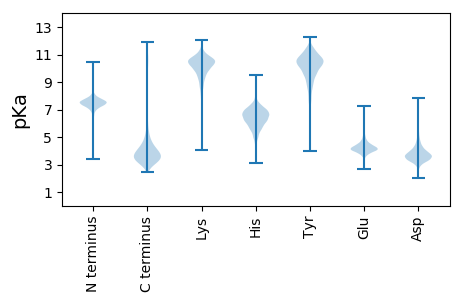
MM1 pKa = 7.2FGVASFYY8 pKa = 11.38AVLGVPMSARR18 pKa = 11.84SWLITSALTVAAVTLVGCAQPTPEE42 pKa = 4.34PSVTPVAEE50 pKa = 4.15PVHH53 pKa = 5.92WSYY56 pKa = 11.99DD57 pKa = 3.64GASGPASWAEE67 pKa = 3.63LDD69 pKa = 4.82SSFEE73 pKa = 4.15TCAVGTDD80 pKa = 3.6QSPIDD85 pKa = 4.2LPATVPAPSTSIRR98 pKa = 11.84LSAEE102 pKa = 3.96DD103 pKa = 3.77ADD105 pKa = 4.37GDD107 pKa = 4.47VFDD110 pKa = 4.9TGHH113 pKa = 6.2AVEE116 pKa = 5.24FEE118 pKa = 4.16SDD120 pKa = 3.8GEE122 pKa = 4.59GEE124 pKa = 4.43TLTFAGDD131 pKa = 3.83EE132 pKa = 4.23YY133 pKa = 11.5SLQQMHH139 pKa = 6.24AHH141 pKa = 5.95VPSEE145 pKa = 4.14HH146 pKa = 5.99TVNGQPAAAEE156 pKa = 3.99LHH158 pKa = 6.39LVHH161 pKa = 7.19ADD163 pKa = 3.0ADD165 pKa = 3.86GRR167 pKa = 11.84LLVLGILVTEE177 pKa = 5.03GQASDD182 pKa = 3.48ALMPFIEE189 pKa = 4.85AATHH193 pKa = 5.61VADD196 pKa = 4.53EE197 pKa = 4.81EE198 pKa = 4.5DD199 pKa = 3.37VTMDD203 pKa = 3.17VSAVLPPSLEE213 pKa = 3.94NYY215 pKa = 8.29EE216 pKa = 4.14YY217 pKa = 10.78SGSLTTPPCTEE228 pKa = 3.83DD229 pKa = 3.57VQWVVMSTPISMSAEE244 pKa = 3.82QIGTLEE250 pKa = 4.49GAHH253 pKa = 5.85SHH255 pKa = 5.76NARR258 pKa = 11.84PTQPLGDD265 pKa = 3.37RR266 pKa = 11.84VVVGGTADD274 pKa = 4.98LDD276 pKa = 3.98VEE278 pKa = 4.54DD279 pKa = 4.51
MM1 pKa = 7.2FGVASFYY8 pKa = 11.38AVLGVPMSARR18 pKa = 11.84SWLITSALTVAAVTLVGCAQPTPEE42 pKa = 4.34PSVTPVAEE50 pKa = 4.15PVHH53 pKa = 5.92WSYY56 pKa = 11.99DD57 pKa = 3.64GASGPASWAEE67 pKa = 3.63LDD69 pKa = 4.82SSFEE73 pKa = 4.15TCAVGTDD80 pKa = 3.6QSPIDD85 pKa = 4.2LPATVPAPSTSIRR98 pKa = 11.84LSAEE102 pKa = 3.96DD103 pKa = 3.77ADD105 pKa = 4.37GDD107 pKa = 4.47VFDD110 pKa = 4.9TGHH113 pKa = 6.2AVEE116 pKa = 5.24FEE118 pKa = 4.16SDD120 pKa = 3.8GEE122 pKa = 4.59GEE124 pKa = 4.43TLTFAGDD131 pKa = 3.83EE132 pKa = 4.23YY133 pKa = 11.5SLQQMHH139 pKa = 6.24AHH141 pKa = 5.95VPSEE145 pKa = 4.14HH146 pKa = 5.99TVNGQPAAAEE156 pKa = 3.99LHH158 pKa = 6.39LVHH161 pKa = 7.19ADD163 pKa = 3.0ADD165 pKa = 3.86GRR167 pKa = 11.84LLVLGILVTEE177 pKa = 5.03GQASDD182 pKa = 3.48ALMPFIEE189 pKa = 4.85AATHH193 pKa = 5.61VADD196 pKa = 4.53EE197 pKa = 4.81EE198 pKa = 4.5DD199 pKa = 3.37VTMDD203 pKa = 3.17VSAVLPPSLEE213 pKa = 3.94NYY215 pKa = 8.29EE216 pKa = 4.14YY217 pKa = 10.78SGSLTTPPCTEE228 pKa = 3.83DD229 pKa = 3.57VQWVVMSTPISMSAEE244 pKa = 3.82QIGTLEE250 pKa = 4.49GAHH253 pKa = 5.85SHH255 pKa = 5.76NARR258 pKa = 11.84PTQPLGDD265 pKa = 3.37RR266 pKa = 11.84VVVGGTADD274 pKa = 4.98LDD276 pKa = 3.98VEE278 pKa = 4.54DD279 pKa = 4.51
Molecular weight: 29.14 kDa
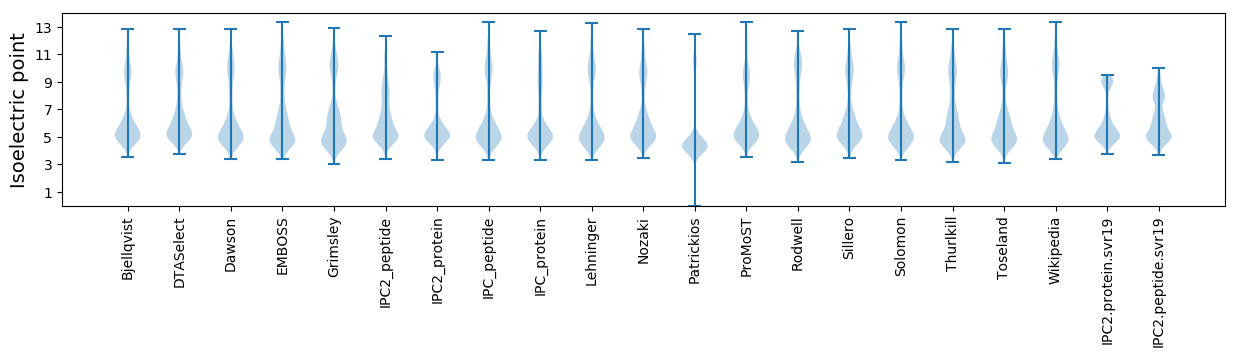
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R9B4C3|A0A4R9B4C3_9MICO CbiA domain-containing protein OS=Cryobacterium sp. Hh4 OX=1259157 GN=E3T48_11555 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
988464 |
30 |
1966 |
315.8 |
33.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.127 ± 0.06 | 0.558 ± 0.011 |
5.868 ± 0.036 | 5.211 ± 0.047 |
3.244 ± 0.029 | 9.052 ± 0.042 |
1.966 ± 0.019 | 4.708 ± 0.035 |
2.25 ± 0.034 | 10.513 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.806 ± 0.016 | 2.241 ± 0.023 |
5.367 ± 0.033 | 2.805 ± 0.023 |
7.058 ± 0.049 | 5.829 ± 0.027 |
6.201 ± 0.038 | 8.783 ± 0.044 |
1.407 ± 0.016 | 2.006 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
