
Komagataella phaffii (strain GS115 / ATCC 20864) (Yeast) (Pichia pastoris)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Phaffomycetaceae; Komagataella; Komagataella phaffii
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
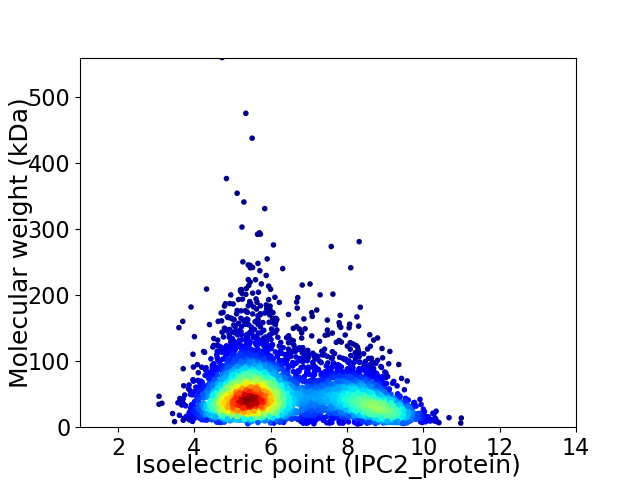
Virtual 2D-PAGE plot for 5073 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C4QWT3|C4QWT3_KOMPG MICOS complex subunit MIC10 OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=PAS_chr1-1_0330 PE=3 SV=1
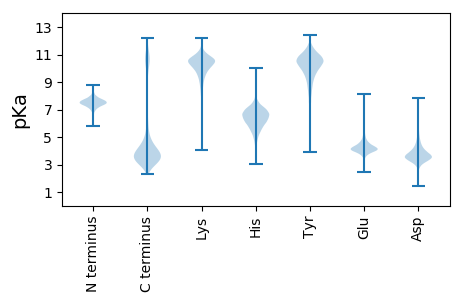
MM1 pKa = 7.65RR2 pKa = 11.84PVLSLLLLLASSVLADD18 pKa = 4.04EE19 pKa = 5.8VIEE22 pKa = 4.41CDD24 pKa = 4.71ADD26 pKa = 3.85NKK28 pKa = 10.72CPEE31 pKa = 4.95DD32 pKa = 4.31KK33 pKa = 10.34PCCSQYY39 pKa = 10.84GVCGTGVNCLGGCDD53 pKa = 3.84PRR55 pKa = 11.84HH56 pKa = 6.08SFNASACLPMPVCRR70 pKa = 11.84DD71 pKa = 3.06VDD73 pKa = 4.42LKK75 pKa = 11.46ASTDD79 pKa = 3.21AFEE82 pKa = 5.9IDD84 pKa = 3.38TNYY87 pKa = 10.9LGDD90 pKa = 4.05ANEE93 pKa = 4.28TDD95 pKa = 3.28WVYY98 pKa = 11.44NGYY101 pKa = 10.47LIDD104 pKa = 4.35YY105 pKa = 9.32DD106 pKa = 5.01DD107 pKa = 4.52SVLLAMPKK115 pKa = 9.77EE116 pKa = 4.5SYY118 pKa = 9.18GTVVSSTFYY127 pKa = 10.15VWYY130 pKa = 10.08GKK132 pKa = 8.1ITATLKK138 pKa = 8.57TSRR141 pKa = 11.84GAGVVTSFILFSNVHH156 pKa = 6.91DD157 pKa = 5.0EE158 pKa = 4.19IDD160 pKa = 3.59WEE162 pKa = 4.36FVGYY166 pKa = 10.18NLSQVEE172 pKa = 4.29TNYY175 pKa = 10.72YY176 pKa = 8.05YY177 pKa = 10.54QGVLNYY183 pKa = 9.97TNGRR187 pKa = 11.84NVSLEE192 pKa = 3.69EE193 pKa = 4.02DD194 pKa = 3.86VNSFEE199 pKa = 4.33YY200 pKa = 10.41FHH202 pKa = 8.04DD203 pKa = 4.56YY204 pKa = 10.77EE205 pKa = 5.6IDD207 pKa = 3.26WKK209 pKa = 10.87EE210 pKa = 4.64DD211 pKa = 3.41VITWSIDD218 pKa = 2.88GDD220 pKa = 3.95VVRR223 pKa = 11.84TLKK226 pKa = 10.95KK227 pKa = 9.81EE228 pKa = 3.72DD229 pKa = 3.82TYY231 pKa = 11.96NEE233 pKa = 4.18TTDD236 pKa = 4.49KK237 pKa = 11.47YY238 pKa = 10.19MFPQTPSRR246 pKa = 11.84VQLSIWPAGAEE257 pKa = 4.13SNAIGTVSWAGGNVDD272 pKa = 4.36WDD274 pKa = 4.0SEE276 pKa = 4.55DD277 pKa = 3.48IQDD280 pKa = 4.11PGYY283 pKa = 10.54FYY285 pKa = 10.88YY286 pKa = 10.13TLKK289 pKa = 10.62EE290 pKa = 4.14LTVEE294 pKa = 4.62CYY296 pKa = 10.07DD297 pKa = 3.73VPDD300 pKa = 3.82GTEE303 pKa = 3.71EE304 pKa = 4.26DD305 pKa = 4.41GEE307 pKa = 3.97LAYY310 pKa = 10.75YY311 pKa = 10.35FKK313 pKa = 11.03EE314 pKa = 4.06SDD316 pKa = 4.03AFDD319 pKa = 3.34QGDD322 pKa = 3.8IIITNNSTKK331 pKa = 10.16IKK333 pKa = 10.71SLDD336 pKa = 3.52DD337 pKa = 3.33TGFDD341 pKa = 4.43PDD343 pKa = 4.8EE344 pKa = 6.41DD345 pKa = 5.63DD346 pKa = 6.5DD347 pKa = 6.26DD348 pKa = 6.44DD349 pKa = 4.85EE350 pKa = 5.94SSSSSSSSSRR360 pKa = 11.84SSSSSSRR367 pKa = 11.84TGSSSTSSATSTSTSNSNDD386 pKa = 3.3DD387 pKa = 4.24DD388 pKa = 5.07DD389 pKa = 5.66NNNSSPTTSSGTSSAASGFVQNMSQTSGSSSATSNNAAASLSAGFLTTISFFASVLGFLL448 pKa = 4.84
MM1 pKa = 7.65RR2 pKa = 11.84PVLSLLLLLASSVLADD18 pKa = 4.04EE19 pKa = 5.8VIEE22 pKa = 4.41CDD24 pKa = 4.71ADD26 pKa = 3.85NKK28 pKa = 10.72CPEE31 pKa = 4.95DD32 pKa = 4.31KK33 pKa = 10.34PCCSQYY39 pKa = 10.84GVCGTGVNCLGGCDD53 pKa = 3.84PRR55 pKa = 11.84HH56 pKa = 6.08SFNASACLPMPVCRR70 pKa = 11.84DD71 pKa = 3.06VDD73 pKa = 4.42LKK75 pKa = 11.46ASTDD79 pKa = 3.21AFEE82 pKa = 5.9IDD84 pKa = 3.38TNYY87 pKa = 10.9LGDD90 pKa = 4.05ANEE93 pKa = 4.28TDD95 pKa = 3.28WVYY98 pKa = 11.44NGYY101 pKa = 10.47LIDD104 pKa = 4.35YY105 pKa = 9.32DD106 pKa = 5.01DD107 pKa = 4.52SVLLAMPKK115 pKa = 9.77EE116 pKa = 4.5SYY118 pKa = 9.18GTVVSSTFYY127 pKa = 10.15VWYY130 pKa = 10.08GKK132 pKa = 8.1ITATLKK138 pKa = 8.57TSRR141 pKa = 11.84GAGVVTSFILFSNVHH156 pKa = 6.91DD157 pKa = 5.0EE158 pKa = 4.19IDD160 pKa = 3.59WEE162 pKa = 4.36FVGYY166 pKa = 10.18NLSQVEE172 pKa = 4.29TNYY175 pKa = 10.72YY176 pKa = 8.05YY177 pKa = 10.54QGVLNYY183 pKa = 9.97TNGRR187 pKa = 11.84NVSLEE192 pKa = 3.69EE193 pKa = 4.02DD194 pKa = 3.86VNSFEE199 pKa = 4.33YY200 pKa = 10.41FHH202 pKa = 8.04DD203 pKa = 4.56YY204 pKa = 10.77EE205 pKa = 5.6IDD207 pKa = 3.26WKK209 pKa = 10.87EE210 pKa = 4.64DD211 pKa = 3.41VITWSIDD218 pKa = 2.88GDD220 pKa = 3.95VVRR223 pKa = 11.84TLKK226 pKa = 10.95KK227 pKa = 9.81EE228 pKa = 3.72DD229 pKa = 3.82TYY231 pKa = 11.96NEE233 pKa = 4.18TTDD236 pKa = 4.49KK237 pKa = 11.47YY238 pKa = 10.19MFPQTPSRR246 pKa = 11.84VQLSIWPAGAEE257 pKa = 4.13SNAIGTVSWAGGNVDD272 pKa = 4.36WDD274 pKa = 4.0SEE276 pKa = 4.55DD277 pKa = 3.48IQDD280 pKa = 4.11PGYY283 pKa = 10.54FYY285 pKa = 10.88YY286 pKa = 10.13TLKK289 pKa = 10.62EE290 pKa = 4.14LTVEE294 pKa = 4.62CYY296 pKa = 10.07DD297 pKa = 3.73VPDD300 pKa = 3.82GTEE303 pKa = 3.71EE304 pKa = 4.26DD305 pKa = 4.41GEE307 pKa = 3.97LAYY310 pKa = 10.75YY311 pKa = 10.35FKK313 pKa = 11.03EE314 pKa = 4.06SDD316 pKa = 4.03AFDD319 pKa = 3.34QGDD322 pKa = 3.8IIITNNSTKK331 pKa = 10.16IKK333 pKa = 10.71SLDD336 pKa = 3.52DD337 pKa = 3.33TGFDD341 pKa = 4.43PDD343 pKa = 4.8EE344 pKa = 6.41DD345 pKa = 5.63DD346 pKa = 6.5DD347 pKa = 6.26DD348 pKa = 6.44DD349 pKa = 4.85EE350 pKa = 5.94SSSSSSSSSRR360 pKa = 11.84SSSSSSRR367 pKa = 11.84TGSSSTSSATSTSTSNSNDD386 pKa = 3.3DD387 pKa = 4.24DD388 pKa = 5.07DD389 pKa = 5.66NNNSSPTTSSGTSSAASGFVQNMSQTSGSSSATSNNAAASLSAGFLTTISFFASVLGFLL448 pKa = 4.84
Molecular weight: 48.77 kDa
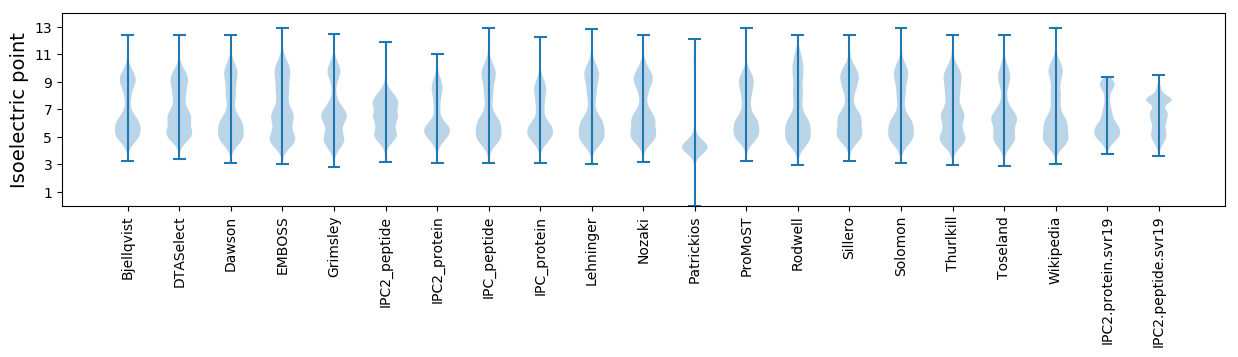
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C4R3E9|C4R3E9_KOMPG Mitochondrial intermembrane space cysteine motif protein OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=PAS_chr3_0056 PE=3 SV=1
MM1 pKa = 7.27PCHH4 pKa = 6.55RR5 pKa = 11.84RR6 pKa = 11.84MMDD9 pKa = 2.5IRR11 pKa = 11.84KK12 pKa = 9.4VIKK15 pKa = 10.31FSDD18 pKa = 3.44VGDD21 pKa = 3.2ITMAVCGIHH30 pKa = 5.83FRR32 pKa = 11.84CRR34 pKa = 11.84LFSFNKK40 pKa = 9.42GIRR43 pKa = 11.84RR44 pKa = 11.84QSYY47 pKa = 7.78STII50 pKa = 3.5
MM1 pKa = 7.27PCHH4 pKa = 6.55RR5 pKa = 11.84RR6 pKa = 11.84MMDD9 pKa = 2.5IRR11 pKa = 11.84KK12 pKa = 9.4VIKK15 pKa = 10.31FSDD18 pKa = 3.44VGDD21 pKa = 3.2ITMAVCGIHH30 pKa = 5.83FRR32 pKa = 11.84CRR34 pKa = 11.84LFSFNKK40 pKa = 9.42GIRR43 pKa = 11.84RR44 pKa = 11.84QSYY47 pKa = 7.78STII50 pKa = 3.5
Molecular weight: 5.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2416083 |
45 |
4950 |
476.3 |
53.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.514 ± 0.027 | 1.17 ± 0.013 |
5.917 ± 0.024 | 6.786 ± 0.037 |
4.463 ± 0.023 | 5.127 ± 0.031 |
2.13 ± 0.014 | 6.34 ± 0.025 |
6.944 ± 0.03 | 10.131 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.853 ± 0.012 | 5.34 ± 0.023 |
4.489 ± 0.025 | 4.193 ± 0.029 |
4.554 ± 0.019 | 8.916 ± 0.041 |
5.69 ± 0.028 | 6.069 ± 0.023 |
1.035 ± 0.011 | 3.333 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
