
Oscillatoriales cyanobacterium USR001
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Oscillatoriales; unclassified Oscillatoriales
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
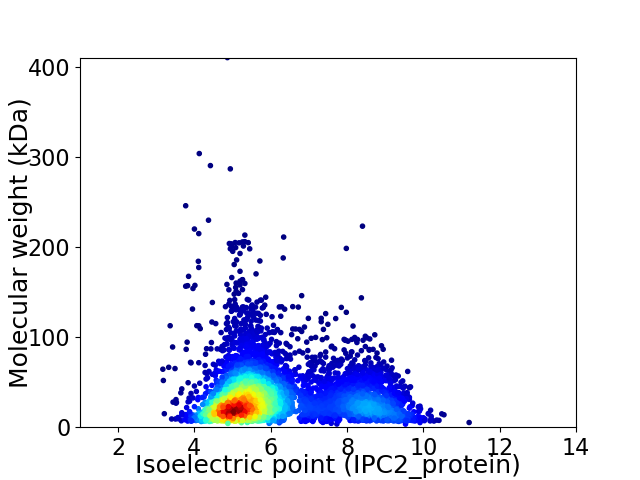
Virtual 2D-PAGE plot for 4908 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1C0VT21|A0A1C0VT21_9CYAN DUF2281 domain-containing protein OS=Oscillatoriales cyanobacterium USR001 OX=1880991 GN=BCD67_02985 PE=4 SV=1
MM1 pKa = 7.72SGGLFGWIGDD11 pKa = 4.02GLAEE15 pKa = 4.32IASEE19 pKa = 4.27VIGVFKK25 pKa = 10.95SDD27 pKa = 4.26DD28 pKa = 3.63ASGSNPGATQGTTTSRR44 pKa = 11.84NQEE47 pKa = 3.92RR48 pKa = 11.84GRR50 pKa = 11.84ASYY53 pKa = 10.54LQGQDD58 pKa = 3.1LEE60 pKa = 5.4RR61 pKa = 11.84YY62 pKa = 7.93TEE64 pKa = 3.88QQLDD68 pKa = 3.92EE69 pKa = 5.07FGLNALWDD77 pKa = 4.16KK78 pKa = 11.62NNLVYY83 pKa = 10.86DD84 pKa = 4.17EE85 pKa = 4.76TLEE88 pKa = 4.15QYY90 pKa = 11.37GKK92 pKa = 6.96TTQVDD97 pKa = 3.52VLIRR101 pKa = 11.84DD102 pKa = 3.84GNDD105 pKa = 2.91NEE107 pKa = 4.38VAIIEE112 pKa = 4.51CKK114 pKa = 10.47AYY116 pKa = 8.78TSKK119 pKa = 10.73WNDD122 pKa = 3.27DD123 pKa = 3.82DD124 pKa = 6.13AVDD127 pKa = 3.45QARR130 pKa = 11.84RR131 pKa = 11.84LVSLANEE138 pKa = 4.04RR139 pKa = 11.84GIPLIFSTYY148 pKa = 10.93DD149 pKa = 2.93GTTDD153 pKa = 3.48CFGSRR158 pKa = 11.84VKK160 pKa = 10.58EE161 pKa = 4.02VLNQTQYY168 pKa = 11.4FVISSDD174 pKa = 3.66NIFFADD180 pKa = 3.77PSDD183 pKa = 3.64VKK185 pKa = 11.25EE186 pKa = 4.24NGVTQGLPPEE196 pKa = 4.93LRR198 pKa = 11.84SHH200 pKa = 6.36WDD202 pKa = 2.21WWFGQNYY209 pKa = 7.84EE210 pKa = 4.58PKK212 pKa = 9.85SSCRR216 pKa = 11.84SDD218 pKa = 3.64YY219 pKa = 11.4SSNDD223 pKa = 3.03PNGFDD228 pKa = 3.37SSYY231 pKa = 8.76EE232 pKa = 4.23TEE234 pKa = 4.47SSSSPDD240 pKa = 3.38YY241 pKa = 10.79EE242 pKa = 4.84RR243 pKa = 11.84SDD245 pKa = 3.76SADD248 pKa = 3.36SGGSWDD254 pKa = 5.58LFSNWFGDD262 pKa = 3.48SSEE265 pKa = 4.89AEE267 pKa = 3.98SSNNSNSSSINSRR280 pKa = 11.84DD281 pKa = 3.46SGNSWFSNWFDD292 pKa = 3.98DD293 pKa = 4.08SSEE296 pKa = 4.43SEE298 pKa = 4.22SSNASDD304 pKa = 3.77SSSINSNSRR313 pKa = 11.84DD314 pKa = 3.41SSDD317 pKa = 3.84YY318 pKa = 10.9GSSDD322 pKa = 3.69YY323 pKa = 11.64GSGDD327 pKa = 3.43YY328 pKa = 11.3GNSGDD333 pKa = 3.78SDD335 pKa = 4.29YY336 pKa = 11.71GSSHH340 pKa = 6.43YY341 pKa = 11.04GSGDD345 pKa = 3.39YY346 pKa = 11.0GDD348 pKa = 5.4SGDD351 pKa = 4.22SDD353 pKa = 4.2SGSSSSDD360 pKa = 3.19SGDD363 pKa = 3.21SDD365 pKa = 4.2YY366 pKa = 11.74GSSDD370 pKa = 3.75YY371 pKa = 11.61GSGDD375 pKa = 3.59YY376 pKa = 11.04GDD378 pKa = 4.93SSGSDD383 pKa = 3.36SSSSSSDD390 pKa = 3.22SWWW393 pKa = 3.06
MM1 pKa = 7.72SGGLFGWIGDD11 pKa = 4.02GLAEE15 pKa = 4.32IASEE19 pKa = 4.27VIGVFKK25 pKa = 10.95SDD27 pKa = 4.26DD28 pKa = 3.63ASGSNPGATQGTTTSRR44 pKa = 11.84NQEE47 pKa = 3.92RR48 pKa = 11.84GRR50 pKa = 11.84ASYY53 pKa = 10.54LQGQDD58 pKa = 3.1LEE60 pKa = 5.4RR61 pKa = 11.84YY62 pKa = 7.93TEE64 pKa = 3.88QQLDD68 pKa = 3.92EE69 pKa = 5.07FGLNALWDD77 pKa = 4.16KK78 pKa = 11.62NNLVYY83 pKa = 10.86DD84 pKa = 4.17EE85 pKa = 4.76TLEE88 pKa = 4.15QYY90 pKa = 11.37GKK92 pKa = 6.96TTQVDD97 pKa = 3.52VLIRR101 pKa = 11.84DD102 pKa = 3.84GNDD105 pKa = 2.91NEE107 pKa = 4.38VAIIEE112 pKa = 4.51CKK114 pKa = 10.47AYY116 pKa = 8.78TSKK119 pKa = 10.73WNDD122 pKa = 3.27DD123 pKa = 3.82DD124 pKa = 6.13AVDD127 pKa = 3.45QARR130 pKa = 11.84RR131 pKa = 11.84LVSLANEE138 pKa = 4.04RR139 pKa = 11.84GIPLIFSTYY148 pKa = 10.93DD149 pKa = 2.93GTTDD153 pKa = 3.48CFGSRR158 pKa = 11.84VKK160 pKa = 10.58EE161 pKa = 4.02VLNQTQYY168 pKa = 11.4FVISSDD174 pKa = 3.66NIFFADD180 pKa = 3.77PSDD183 pKa = 3.64VKK185 pKa = 11.25EE186 pKa = 4.24NGVTQGLPPEE196 pKa = 4.93LRR198 pKa = 11.84SHH200 pKa = 6.36WDD202 pKa = 2.21WWFGQNYY209 pKa = 7.84EE210 pKa = 4.58PKK212 pKa = 9.85SSCRR216 pKa = 11.84SDD218 pKa = 3.64YY219 pKa = 11.4SSNDD223 pKa = 3.03PNGFDD228 pKa = 3.37SSYY231 pKa = 8.76EE232 pKa = 4.23TEE234 pKa = 4.47SSSSPDD240 pKa = 3.38YY241 pKa = 10.79EE242 pKa = 4.84RR243 pKa = 11.84SDD245 pKa = 3.76SADD248 pKa = 3.36SGGSWDD254 pKa = 5.58LFSNWFGDD262 pKa = 3.48SSEE265 pKa = 4.89AEE267 pKa = 3.98SSNNSNSSSINSRR280 pKa = 11.84DD281 pKa = 3.46SGNSWFSNWFDD292 pKa = 3.98DD293 pKa = 4.08SSEE296 pKa = 4.43SEE298 pKa = 4.22SSNASDD304 pKa = 3.77SSSINSNSRR313 pKa = 11.84DD314 pKa = 3.41SSDD317 pKa = 3.84YY318 pKa = 10.9GSSDD322 pKa = 3.69YY323 pKa = 11.64GSGDD327 pKa = 3.43YY328 pKa = 11.3GNSGDD333 pKa = 3.78SDD335 pKa = 4.29YY336 pKa = 11.71GSSHH340 pKa = 6.43YY341 pKa = 11.04GSGDD345 pKa = 3.39YY346 pKa = 11.0GDD348 pKa = 5.4SGDD351 pKa = 4.22SDD353 pKa = 4.2SGSSSSDD360 pKa = 3.19SGDD363 pKa = 3.21SDD365 pKa = 4.2YY366 pKa = 11.74GSSDD370 pKa = 3.75YY371 pKa = 11.61GSGDD375 pKa = 3.59YY376 pKa = 11.04GDD378 pKa = 4.93SSGSDD383 pKa = 3.36SSSSSSDD390 pKa = 3.22SWWW393 pKa = 3.06
Molecular weight: 42.55 kDa
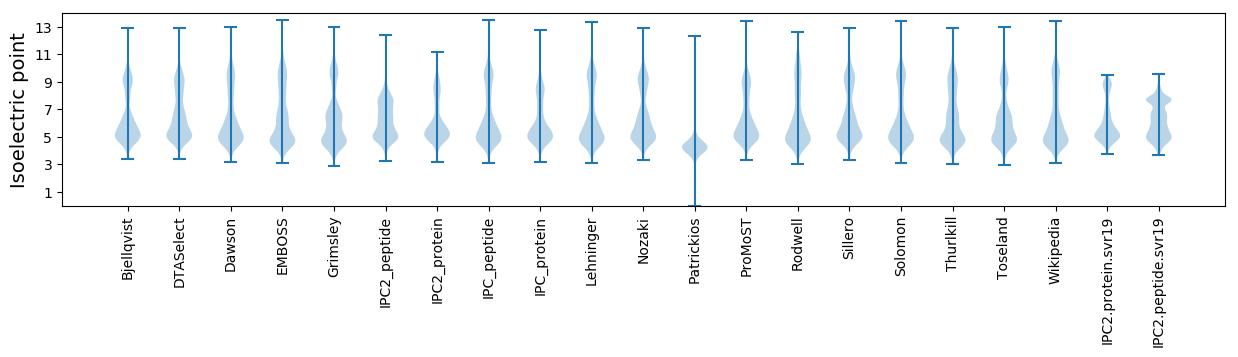
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1C0VW16|A0A1C0VW16_9CYAN Cytosine-specific methyltransferase OS=Oscillatoriales cyanobacterium USR001 OX=1880991 GN=BCD67_05635 PE=3 SV=1
MM1 pKa = 6.86TQQTLHH7 pKa = 5.03GTSRR11 pKa = 11.84KK12 pKa = 9.46RR13 pKa = 11.84KK14 pKa = 7.05RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.26SGQAVISARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05KK38 pKa = 10.18GRR40 pKa = 11.84HH41 pKa = 4.82RR42 pKa = 11.84LAVV45 pKa = 3.37
MM1 pKa = 6.86TQQTLHH7 pKa = 5.03GTSRR11 pKa = 11.84KK12 pKa = 9.46RR13 pKa = 11.84KK14 pKa = 7.05RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.26SGQAVISARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05KK38 pKa = 10.18GRR40 pKa = 11.84HH41 pKa = 4.82RR42 pKa = 11.84LAVV45 pKa = 3.37
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1600603 |
31 |
3712 |
326.1 |
36.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.839 ± 0.037 | 0.969 ± 0.013 |
4.959 ± 0.027 | 6.709 ± 0.04 |
3.969 ± 0.022 | 6.672 ± 0.049 |
1.609 ± 0.018 | 7.32 ± 0.033 |
5.207 ± 0.036 | 10.888 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.884 ± 0.016 | 4.768 ± 0.035 |
4.646 ± 0.028 | 5.015 ± 0.035 |
5.006 ± 0.031 | 6.435 ± 0.036 |
5.543 ± 0.039 | 6.206 ± 0.029 |
1.366 ± 0.017 | 2.992 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
