
CRESS virus sp. ctmn412
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; unclassified Cressdnaviricota; CRESS viruses
Average proteome isoelectric point is 8.3
Get precalculated fractions of proteins
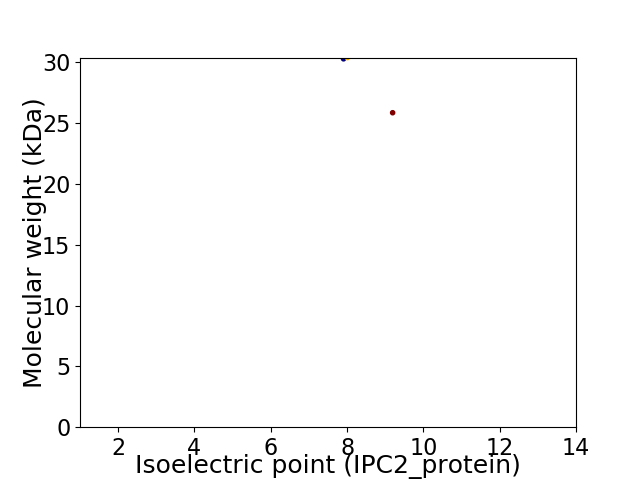
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
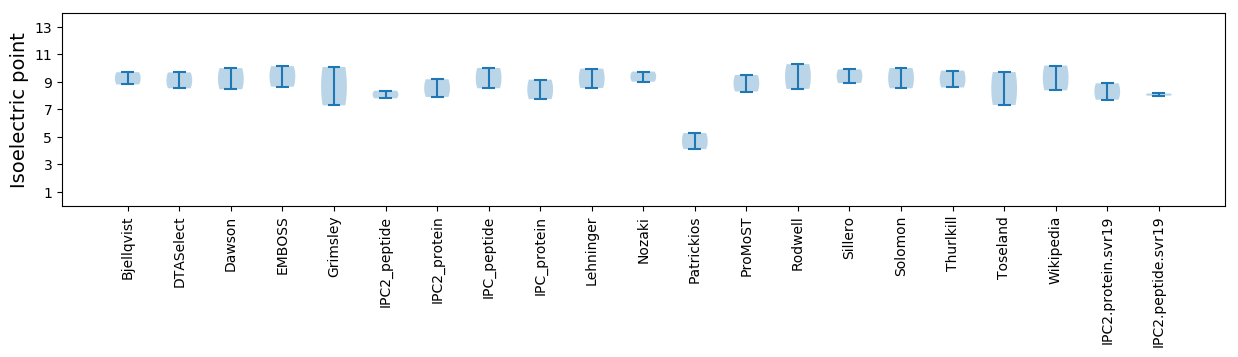
Summary statistics related to proteome-wise predictions
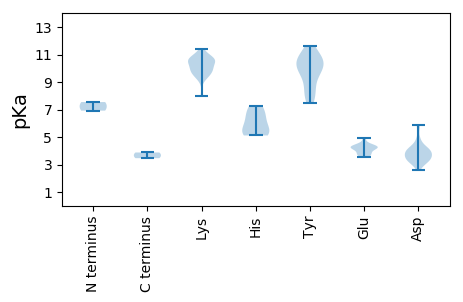
Protein with the lowest isoelectric point:
>tr|A0A5Q2W5N0|A0A5Q2W5N0_9VIRU Rep protein OS=CRESS virus sp. ctmn412 OX=2656688 PE=4 SV=1
MM1 pKa = 7.54ARR3 pKa = 11.84QSNSKK8 pKa = 9.66KK9 pKa = 9.5PRR11 pKa = 11.84VNRR14 pKa = 11.84SRR16 pKa = 11.84SWFFTYY22 pKa = 10.82NNPPEE27 pKa = 4.32NAWAQIIDD35 pKa = 3.88TFEE38 pKa = 4.19NDD40 pKa = 2.63QCEE43 pKa = 4.21YY44 pKa = 11.09VFQLEE49 pKa = 4.29QGDD52 pKa = 4.16TVHH55 pKa = 5.77YY56 pKa = 10.08QGVIRR61 pKa = 11.84YY62 pKa = 7.48KK63 pKa = 10.57NPRR66 pKa = 11.84EE67 pKa = 3.68NWPQISCHH75 pKa = 5.35WEE77 pKa = 3.71RR78 pKa = 11.84CISWRR83 pKa = 11.84SAIKK87 pKa = 10.51YY88 pKa = 8.62CSKK91 pKa = 9.91VQTRR95 pKa = 11.84IDD97 pKa = 4.11GPWTNIDD104 pKa = 3.36GLKK107 pKa = 10.55FRR109 pKa = 11.84DD110 pKa = 4.06TIKK113 pKa = 11.15DD114 pKa = 3.28PLQNKK119 pKa = 9.1KK120 pKa = 10.47LYY122 pKa = 9.83PFQQEE127 pKa = 3.57ILDD130 pKa = 4.27IIKK133 pKa = 9.59TDD135 pKa = 3.34SNDD138 pKa = 3.82RR139 pKa = 11.84VIYY142 pKa = 8.88WYY144 pKa = 9.54WEE146 pKa = 4.01PNGNTGKK153 pKa = 10.69SSLCKK158 pKa = 10.22HH159 pKa = 5.25IMLRR163 pKa = 11.84YY164 pKa = 8.15KK165 pKa = 10.54ACLIGGAAKK174 pKa = 10.77DD175 pKa = 3.62NFHH178 pKa = 6.74CVSSYY183 pKa = 11.64LEE185 pKa = 4.91DD186 pKa = 3.98NDD188 pKa = 5.73LKK190 pKa = 11.39VCLIDD195 pKa = 4.81IPRR198 pKa = 11.84CSMNSVSYY206 pKa = 8.56KK207 pKa = 10.04TIEE210 pKa = 4.47CIKK213 pKa = 10.7NGIIFSGKK221 pKa = 9.63YY222 pKa = 7.86EE223 pKa = 4.37SNYY226 pKa = 10.62KK227 pKa = 9.99IFNSPHH233 pKa = 7.25LIILANFPPNTDD245 pKa = 3.23MLSEE249 pKa = 4.32DD250 pKa = 2.89RR251 pKa = 11.84WVVKK255 pKa = 10.53RR256 pKa = 11.84II257 pKa = 3.46
MM1 pKa = 7.54ARR3 pKa = 11.84QSNSKK8 pKa = 9.66KK9 pKa = 9.5PRR11 pKa = 11.84VNRR14 pKa = 11.84SRR16 pKa = 11.84SWFFTYY22 pKa = 10.82NNPPEE27 pKa = 4.32NAWAQIIDD35 pKa = 3.88TFEE38 pKa = 4.19NDD40 pKa = 2.63QCEE43 pKa = 4.21YY44 pKa = 11.09VFQLEE49 pKa = 4.29QGDD52 pKa = 4.16TVHH55 pKa = 5.77YY56 pKa = 10.08QGVIRR61 pKa = 11.84YY62 pKa = 7.48KK63 pKa = 10.57NPRR66 pKa = 11.84EE67 pKa = 3.68NWPQISCHH75 pKa = 5.35WEE77 pKa = 3.71RR78 pKa = 11.84CISWRR83 pKa = 11.84SAIKK87 pKa = 10.51YY88 pKa = 8.62CSKK91 pKa = 9.91VQTRR95 pKa = 11.84IDD97 pKa = 4.11GPWTNIDD104 pKa = 3.36GLKK107 pKa = 10.55FRR109 pKa = 11.84DD110 pKa = 4.06TIKK113 pKa = 11.15DD114 pKa = 3.28PLQNKK119 pKa = 9.1KK120 pKa = 10.47LYY122 pKa = 9.83PFQQEE127 pKa = 3.57ILDD130 pKa = 4.27IIKK133 pKa = 9.59TDD135 pKa = 3.34SNDD138 pKa = 3.82RR139 pKa = 11.84VIYY142 pKa = 8.88WYY144 pKa = 9.54WEE146 pKa = 4.01PNGNTGKK153 pKa = 10.69SSLCKK158 pKa = 10.22HH159 pKa = 5.25IMLRR163 pKa = 11.84YY164 pKa = 8.15KK165 pKa = 10.54ACLIGGAAKK174 pKa = 10.77DD175 pKa = 3.62NFHH178 pKa = 6.74CVSSYY183 pKa = 11.64LEE185 pKa = 4.91DD186 pKa = 3.98NDD188 pKa = 5.73LKK190 pKa = 11.39VCLIDD195 pKa = 4.81IPRR198 pKa = 11.84CSMNSVSYY206 pKa = 8.56KK207 pKa = 10.04TIEE210 pKa = 4.47CIKK213 pKa = 10.7NGIIFSGKK221 pKa = 9.63YY222 pKa = 7.86EE223 pKa = 4.37SNYY226 pKa = 10.62KK227 pKa = 9.99IFNSPHH233 pKa = 7.25LIILANFPPNTDD245 pKa = 3.23MLSEE249 pKa = 4.32DD250 pKa = 2.89RR251 pKa = 11.84WVVKK255 pKa = 10.53RR256 pKa = 11.84II257 pKa = 3.46
Molecular weight: 30.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5Q2W5N0|A0A5Q2W5N0_9VIRU Rep protein OS=CRESS virus sp. ctmn412 OX=2656688 PE=4 SV=1
MM1 pKa = 6.92SAYY4 pKa = 10.2RR5 pKa = 11.84GRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.09RR10 pKa = 11.84KK11 pKa = 8.01FGYY14 pKa = 9.38RR15 pKa = 11.84KK16 pKa = 9.55KK17 pKa = 10.3SYY19 pKa = 10.2RR20 pKa = 11.84RR21 pKa = 11.84NAEE24 pKa = 3.87DD25 pKa = 3.75KK26 pKa = 10.69RR27 pKa = 11.84IARR30 pKa = 11.84IAKK33 pKa = 9.87AVTSKK38 pKa = 11.04AKK40 pKa = 9.4EE41 pKa = 3.75LKK43 pKa = 10.66YY44 pKa = 10.77YY45 pKa = 9.69DD46 pKa = 4.14TIITNNTPVSGTSLVSILDD65 pKa = 3.79DD66 pKa = 3.39MAQGDD71 pKa = 4.34TEE73 pKa = 4.27STRR76 pKa = 11.84DD77 pKa = 3.29GNYY80 pKa = 10.02IIIKK84 pKa = 9.2SINVKK89 pKa = 10.46GYY91 pKa = 10.89VYY93 pKa = 10.52DD94 pKa = 3.61QGAGPRR100 pKa = 11.84DD101 pKa = 3.89TICRR105 pKa = 11.84MILIRR110 pKa = 11.84EE111 pKa = 4.16KK112 pKa = 10.86VDD114 pKa = 3.6CEE116 pKa = 4.27GNLPLVGDD124 pKa = 4.83LLDD127 pKa = 3.87TDD129 pKa = 5.87AIPSLRR135 pKa = 11.84YY136 pKa = 7.77WPNMGNFQVWWDD148 pKa = 3.35KK149 pKa = 10.94TFVIRR154 pKa = 11.84ASDD157 pKa = 4.19DD158 pKa = 3.64YY159 pKa = 11.51VTDD162 pKa = 4.03QIQHH166 pKa = 5.11SRR168 pKa = 11.84LVKK171 pKa = 10.07YY172 pKa = 9.69YY173 pKa = 10.51KK174 pKa = 9.95KK175 pKa = 10.41AKK177 pKa = 9.81KK178 pKa = 9.52PIRR181 pKa = 11.84AYY183 pKa = 10.81YY184 pKa = 10.27DD185 pKa = 3.32GVGPGVGTCKK195 pKa = 10.0RR196 pKa = 11.84GHH198 pKa = 6.36FFLITMTDD206 pKa = 3.12QVAGNRR212 pKa = 11.84PNWDD216 pKa = 3.28LKK218 pKa = 10.8VRR220 pKa = 11.84ITFTEE225 pKa = 3.88
MM1 pKa = 6.92SAYY4 pKa = 10.2RR5 pKa = 11.84GRR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 10.09RR10 pKa = 11.84KK11 pKa = 8.01FGYY14 pKa = 9.38RR15 pKa = 11.84KK16 pKa = 9.55KK17 pKa = 10.3SYY19 pKa = 10.2RR20 pKa = 11.84RR21 pKa = 11.84NAEE24 pKa = 3.87DD25 pKa = 3.75KK26 pKa = 10.69RR27 pKa = 11.84IARR30 pKa = 11.84IAKK33 pKa = 9.87AVTSKK38 pKa = 11.04AKK40 pKa = 9.4EE41 pKa = 3.75LKK43 pKa = 10.66YY44 pKa = 10.77YY45 pKa = 9.69DD46 pKa = 4.14TIITNNTPVSGTSLVSILDD65 pKa = 3.79DD66 pKa = 3.39MAQGDD71 pKa = 4.34TEE73 pKa = 4.27STRR76 pKa = 11.84DD77 pKa = 3.29GNYY80 pKa = 10.02IIIKK84 pKa = 9.2SINVKK89 pKa = 10.46GYY91 pKa = 10.89VYY93 pKa = 10.52DD94 pKa = 3.61QGAGPRR100 pKa = 11.84DD101 pKa = 3.89TICRR105 pKa = 11.84MILIRR110 pKa = 11.84EE111 pKa = 4.16KK112 pKa = 10.86VDD114 pKa = 3.6CEE116 pKa = 4.27GNLPLVGDD124 pKa = 4.83LLDD127 pKa = 3.87TDD129 pKa = 5.87AIPSLRR135 pKa = 11.84YY136 pKa = 7.77WPNMGNFQVWWDD148 pKa = 3.35KK149 pKa = 10.94TFVIRR154 pKa = 11.84ASDD157 pKa = 4.19DD158 pKa = 3.64YY159 pKa = 11.51VTDD162 pKa = 4.03QIQHH166 pKa = 5.11SRR168 pKa = 11.84LVKK171 pKa = 10.07YY172 pKa = 9.69YY173 pKa = 10.51KK174 pKa = 9.95KK175 pKa = 10.41AKK177 pKa = 9.81KK178 pKa = 9.52PIRR181 pKa = 11.84AYY183 pKa = 10.81YY184 pKa = 10.27DD185 pKa = 3.32GVGPGVGTCKK195 pKa = 10.0RR196 pKa = 11.84GHH198 pKa = 6.36FFLITMTDD206 pKa = 3.12QVAGNRR212 pKa = 11.84PNWDD216 pKa = 3.28LKK218 pKa = 10.8VRR220 pKa = 11.84ITFTEE225 pKa = 3.88
Molecular weight: 25.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
482 |
225 |
257 |
241.0 |
28.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.357 ± 0.947 | 2.697 ± 0.909 |
7.261 ± 0.788 | 3.734 ± 0.711 |
3.32 ± 0.435 | 5.602 ± 1.302 |
1.452 ± 0.375 | 8.921 ± 0.614 |
8.299 ± 0.393 | 5.394 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.867 ± 0.237 | 6.432 ± 1.324 |
4.357 ± 0.534 | 3.527 ± 0.573 |
7.261 ± 1.084 | 6.432 ± 1.028 |
5.394 ± 1.144 | 5.394 ± 0.848 |
2.697 ± 0.612 | 5.602 ± 0.413 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
