
Capybara microvirus Cap1_SP_107
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
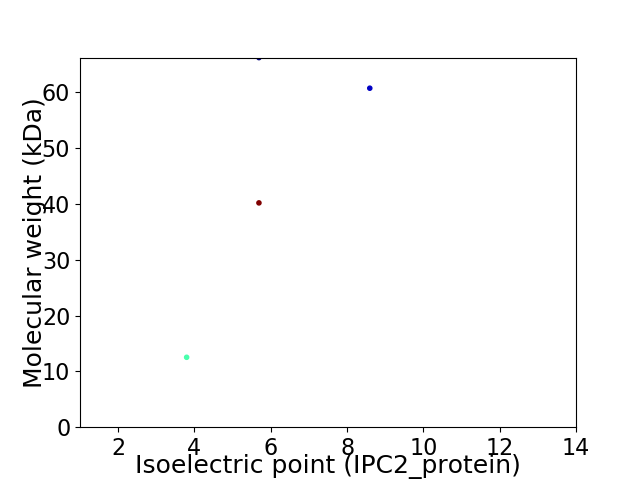
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
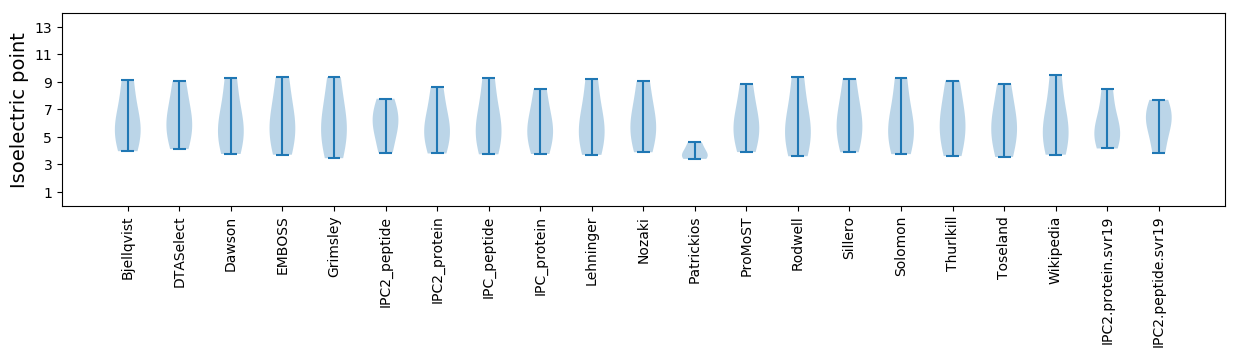
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVN8|A0A4V1FVN8_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_107 OX=2584805 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.74VTSKK6 pKa = 10.62LYY8 pKa = 10.57KK9 pKa = 8.79VGPWFSQPQDD19 pKa = 3.88DD20 pKa = 3.74IRR22 pKa = 11.84VVVAYY27 pKa = 10.36DD28 pKa = 3.48EE29 pKa = 5.89DD30 pKa = 4.48GNQYY34 pKa = 7.61TTFQPYY40 pKa = 10.34EE41 pKa = 4.16GDD43 pKa = 3.3PDD45 pKa = 3.81KK46 pKa = 11.35FGTADD51 pKa = 3.96DD52 pKa = 4.13WSLSNLMAAGIDD64 pKa = 3.37PASFNTTYY72 pKa = 11.2SPLTRR77 pKa = 11.84LDD79 pKa = 3.32SAAEE83 pKa = 3.94FAQISSVAEE92 pKa = 3.83ALIDD96 pKa = 3.94EE97 pKa = 5.15LDD99 pKa = 3.55AAAAEE104 pKa = 4.48PQVSMEE110 pKa = 4.76DD111 pKa = 3.48KK112 pKa = 10.96IEE114 pKa = 3.79
MM1 pKa = 7.44KK2 pKa = 9.74VTSKK6 pKa = 10.62LYY8 pKa = 10.57KK9 pKa = 8.79VGPWFSQPQDD19 pKa = 3.88DD20 pKa = 3.74IRR22 pKa = 11.84VVVAYY27 pKa = 10.36DD28 pKa = 3.48EE29 pKa = 5.89DD30 pKa = 4.48GNQYY34 pKa = 7.61TTFQPYY40 pKa = 10.34EE41 pKa = 4.16GDD43 pKa = 3.3PDD45 pKa = 3.81KK46 pKa = 11.35FGTADD51 pKa = 3.96DD52 pKa = 4.13WSLSNLMAAGIDD64 pKa = 3.37PASFNTTYY72 pKa = 11.2SPLTRR77 pKa = 11.84LDD79 pKa = 3.32SAAEE83 pKa = 3.94FAQISSVAEE92 pKa = 3.83ALIDD96 pKa = 3.94EE97 pKa = 5.15LDD99 pKa = 3.55AAAAEE104 pKa = 4.48PQVSMEE110 pKa = 4.76DD111 pKa = 3.48KK112 pKa = 10.96IEE114 pKa = 3.79
Molecular weight: 12.52 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7S1|A0A4P8W7S1_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_107 OX=2584805 PE=3 SV=1
MM1 pKa = 7.71KK2 pKa = 9.9STITFTNPTTYY13 pKa = 10.47FHH15 pKa = 7.68PDD17 pKa = 3.06YY18 pKa = 10.85SAGSVTIQNPALYY31 pKa = 10.24FSDD34 pKa = 3.37TTKK37 pKa = 10.05QQGYY41 pKa = 5.41EE42 pKa = 3.64TRR44 pKa = 11.84LYY46 pKa = 10.78FEE48 pKa = 4.62YY49 pKa = 10.53EE50 pKa = 3.84GTKK53 pKa = 10.7AKK55 pKa = 10.89GGDD58 pKa = 3.48SYY60 pKa = 11.65FYY62 pKa = 10.75TLTYY66 pKa = 11.07NNAALPHH73 pKa = 6.55YY74 pKa = 10.06IPALNSVVRR83 pKa = 11.84GSQLGGIPCFDD94 pKa = 4.13YY95 pKa = 11.43EE96 pKa = 4.63DD97 pKa = 3.33IRR99 pKa = 11.84YY100 pKa = 7.04ITHH103 pKa = 6.78GGLDD107 pKa = 3.36KK108 pKa = 11.1KK109 pKa = 10.59LLRR112 pKa = 11.84NFGTKK117 pKa = 8.06MKK119 pKa = 10.51YY120 pKa = 9.86FVGCEE125 pKa = 3.73LGEE128 pKa = 4.51GKK130 pKa = 10.54GSRR133 pKa = 11.84GAGNNPHH140 pKa = 5.57YY141 pKa = 10.52HH142 pKa = 5.6VLFFLTNAEE151 pKa = 4.14DD152 pKa = 3.33EE153 pKa = 4.58RR154 pKa = 11.84YY155 pKa = 8.55PYY157 pKa = 10.65HH158 pKa = 7.55RR159 pKa = 11.84ILPEE163 pKa = 3.75QFEE166 pKa = 4.68TLIKK170 pKa = 9.83TYY172 pKa = 9.21WQGTSEE178 pKa = 4.65KK179 pKa = 9.53IDD181 pKa = 3.37YY182 pKa = 10.58SKK184 pKa = 11.64AKK186 pKa = 10.35FGIARR191 pKa = 11.84PGNNLGLLDD200 pKa = 4.35GFAGTLYY207 pKa = 10.06CAKK210 pKa = 9.8YY211 pKa = 8.17VCKK214 pKa = 10.37DD215 pKa = 3.27ASVKK219 pKa = 10.12NLEE222 pKa = 4.28RR223 pKa = 11.84KK224 pKa = 9.78VYY226 pKa = 9.98AQLSSILTDD235 pKa = 3.34EE236 pKa = 4.64NNRR239 pKa = 11.84KK240 pKa = 9.32PIEE243 pKa = 4.14KK244 pKa = 9.99RR245 pKa = 11.84LTPSEE250 pKa = 4.18LEE252 pKa = 3.75KK253 pKa = 10.55EE254 pKa = 4.03IRR256 pKa = 11.84SRR258 pKa = 11.84LTDD261 pKa = 3.4FRR263 pKa = 11.84NRR265 pKa = 11.84YY266 pKa = 6.18SAKK269 pKa = 9.39VRR271 pKa = 11.84ISNGVGNYY279 pKa = 10.57AFIHH283 pKa = 5.62PSFRR287 pKa = 11.84EE288 pKa = 4.05DD289 pKa = 3.24TFSVSMPHH297 pKa = 6.96KK298 pKa = 10.68NGFKK302 pKa = 10.1RR303 pKa = 11.84KK304 pKa = 9.63KK305 pKa = 9.75LGLYY309 pKa = 8.18YY310 pKa = 10.2FRR312 pKa = 11.84KK313 pKa = 8.79YY314 pKa = 9.59FTDD317 pKa = 3.3IVYY320 pKa = 10.55DD321 pKa = 3.74KK322 pKa = 10.45NKK324 pKa = 10.1SPRR327 pKa = 11.84RR328 pKa = 11.84VLNQRR333 pKa = 11.84GIEE336 pKa = 3.9YY337 pKa = 10.26RR338 pKa = 11.84IDD340 pKa = 3.38HH341 pKa = 6.55LGEE344 pKa = 4.15GLEE347 pKa = 4.36KK348 pKa = 10.43KK349 pKa = 10.67AKK351 pKa = 6.4TTYY354 pKa = 10.56KK355 pKa = 10.38CVQFYY360 pKa = 10.86LKK362 pKa = 10.64NSEE365 pKa = 4.04YY366 pKa = 9.98RR367 pKa = 11.84DD368 pKa = 3.54KK369 pKa = 11.58YY370 pKa = 9.24MKK372 pKa = 10.42SPHH375 pKa = 5.69RR376 pKa = 11.84TVEE379 pKa = 3.56PKK381 pKa = 9.44YY382 pKa = 8.97TVKK385 pKa = 10.46EE386 pKa = 4.01LEE388 pKa = 4.16RR389 pKa = 11.84LLSIDD394 pKa = 3.71NNDD397 pKa = 4.58IINSMYY403 pKa = 10.53YY404 pKa = 10.49DD405 pKa = 3.32YY406 pKa = 12.11AMYY409 pKa = 10.79KK410 pKa = 10.21NIYY413 pKa = 7.04EE414 pKa = 3.7HH415 pKa = 6.96HH416 pKa = 6.75YY417 pKa = 7.96YY418 pKa = 10.62YY419 pKa = 10.57EE420 pKa = 4.18YY421 pKa = 10.7TRR423 pKa = 11.84EE424 pKa = 4.16LDD426 pKa = 3.17PMEE429 pKa = 5.04DD430 pKa = 3.17YY431 pKa = 11.17RR432 pKa = 11.84RR433 pKa = 11.84FLTSTFGVDD442 pKa = 3.89GLTTVDD448 pKa = 3.6KK449 pKa = 10.06TALPSGYY456 pKa = 9.54YY457 pKa = 9.78SYY459 pKa = 11.29SRR461 pKa = 11.84HH462 pKa = 5.96PFFNEE467 pKa = 3.71CTDD470 pKa = 3.96LFSLLDD476 pKa = 4.16DD477 pKa = 4.17LVSFYY482 pKa = 10.8FVEE485 pKa = 5.0LSEE488 pKa = 4.41KK489 pKa = 7.17TQKK492 pKa = 10.83DD493 pKa = 3.46YY494 pKa = 11.8EE495 pKa = 4.2EE496 pKa = 4.3RR497 pKa = 11.84KK498 pKa = 8.32RR499 pKa = 11.84TKK501 pKa = 10.71NMYY504 pKa = 10.08RR505 pKa = 11.84DD506 pKa = 3.31LKK508 pKa = 10.62LRR510 pKa = 11.84KK511 pKa = 9.11EE512 pKa = 4.2GLFAVV517 pKa = 4.53
MM1 pKa = 7.71KK2 pKa = 9.9STITFTNPTTYY13 pKa = 10.47FHH15 pKa = 7.68PDD17 pKa = 3.06YY18 pKa = 10.85SAGSVTIQNPALYY31 pKa = 10.24FSDD34 pKa = 3.37TTKK37 pKa = 10.05QQGYY41 pKa = 5.41EE42 pKa = 3.64TRR44 pKa = 11.84LYY46 pKa = 10.78FEE48 pKa = 4.62YY49 pKa = 10.53EE50 pKa = 3.84GTKK53 pKa = 10.7AKK55 pKa = 10.89GGDD58 pKa = 3.48SYY60 pKa = 11.65FYY62 pKa = 10.75TLTYY66 pKa = 11.07NNAALPHH73 pKa = 6.55YY74 pKa = 10.06IPALNSVVRR83 pKa = 11.84GSQLGGIPCFDD94 pKa = 4.13YY95 pKa = 11.43EE96 pKa = 4.63DD97 pKa = 3.33IRR99 pKa = 11.84YY100 pKa = 7.04ITHH103 pKa = 6.78GGLDD107 pKa = 3.36KK108 pKa = 11.1KK109 pKa = 10.59LLRR112 pKa = 11.84NFGTKK117 pKa = 8.06MKK119 pKa = 10.51YY120 pKa = 9.86FVGCEE125 pKa = 3.73LGEE128 pKa = 4.51GKK130 pKa = 10.54GSRR133 pKa = 11.84GAGNNPHH140 pKa = 5.57YY141 pKa = 10.52HH142 pKa = 5.6VLFFLTNAEE151 pKa = 4.14DD152 pKa = 3.33EE153 pKa = 4.58RR154 pKa = 11.84YY155 pKa = 8.55PYY157 pKa = 10.65HH158 pKa = 7.55RR159 pKa = 11.84ILPEE163 pKa = 3.75QFEE166 pKa = 4.68TLIKK170 pKa = 9.83TYY172 pKa = 9.21WQGTSEE178 pKa = 4.65KK179 pKa = 9.53IDD181 pKa = 3.37YY182 pKa = 10.58SKK184 pKa = 11.64AKK186 pKa = 10.35FGIARR191 pKa = 11.84PGNNLGLLDD200 pKa = 4.35GFAGTLYY207 pKa = 10.06CAKK210 pKa = 9.8YY211 pKa = 8.17VCKK214 pKa = 10.37DD215 pKa = 3.27ASVKK219 pKa = 10.12NLEE222 pKa = 4.28RR223 pKa = 11.84KK224 pKa = 9.78VYY226 pKa = 9.98AQLSSILTDD235 pKa = 3.34EE236 pKa = 4.64NNRR239 pKa = 11.84KK240 pKa = 9.32PIEE243 pKa = 4.14KK244 pKa = 9.99RR245 pKa = 11.84LTPSEE250 pKa = 4.18LEE252 pKa = 3.75KK253 pKa = 10.55EE254 pKa = 4.03IRR256 pKa = 11.84SRR258 pKa = 11.84LTDD261 pKa = 3.4FRR263 pKa = 11.84NRR265 pKa = 11.84YY266 pKa = 6.18SAKK269 pKa = 9.39VRR271 pKa = 11.84ISNGVGNYY279 pKa = 10.57AFIHH283 pKa = 5.62PSFRR287 pKa = 11.84EE288 pKa = 4.05DD289 pKa = 3.24TFSVSMPHH297 pKa = 6.96KK298 pKa = 10.68NGFKK302 pKa = 10.1RR303 pKa = 11.84KK304 pKa = 9.63KK305 pKa = 9.75LGLYY309 pKa = 8.18YY310 pKa = 10.2FRR312 pKa = 11.84KK313 pKa = 8.79YY314 pKa = 9.59FTDD317 pKa = 3.3IVYY320 pKa = 10.55DD321 pKa = 3.74KK322 pKa = 10.45NKK324 pKa = 10.1SPRR327 pKa = 11.84RR328 pKa = 11.84VLNQRR333 pKa = 11.84GIEE336 pKa = 3.9YY337 pKa = 10.26RR338 pKa = 11.84IDD340 pKa = 3.38HH341 pKa = 6.55LGEE344 pKa = 4.15GLEE347 pKa = 4.36KK348 pKa = 10.43KK349 pKa = 10.67AKK351 pKa = 6.4TTYY354 pKa = 10.56KK355 pKa = 10.38CVQFYY360 pKa = 10.86LKK362 pKa = 10.64NSEE365 pKa = 4.04YY366 pKa = 9.98RR367 pKa = 11.84DD368 pKa = 3.54KK369 pKa = 11.58YY370 pKa = 9.24MKK372 pKa = 10.42SPHH375 pKa = 5.69RR376 pKa = 11.84TVEE379 pKa = 3.56PKK381 pKa = 9.44YY382 pKa = 8.97TVKK385 pKa = 10.46EE386 pKa = 4.01LEE388 pKa = 4.16RR389 pKa = 11.84LLSIDD394 pKa = 3.71NNDD397 pKa = 4.58IINSMYY403 pKa = 10.53YY404 pKa = 10.49DD405 pKa = 3.32YY406 pKa = 12.11AMYY409 pKa = 10.79KK410 pKa = 10.21NIYY413 pKa = 7.04EE414 pKa = 3.7HH415 pKa = 6.96HH416 pKa = 6.75YY417 pKa = 7.96YY418 pKa = 10.62YY419 pKa = 10.57EE420 pKa = 4.18YY421 pKa = 10.7TRR423 pKa = 11.84EE424 pKa = 4.16LDD426 pKa = 3.17PMEE429 pKa = 5.04DD430 pKa = 3.17YY431 pKa = 11.17RR432 pKa = 11.84RR433 pKa = 11.84FLTSTFGVDD442 pKa = 3.89GLTTVDD448 pKa = 3.6KK449 pKa = 10.06TALPSGYY456 pKa = 9.54YY457 pKa = 9.78SYY459 pKa = 11.29SRR461 pKa = 11.84HH462 pKa = 5.96PFFNEE467 pKa = 3.71CTDD470 pKa = 3.96LFSLLDD476 pKa = 4.16DD477 pKa = 4.17LVSFYY482 pKa = 10.8FVEE485 pKa = 5.0LSEE488 pKa = 4.41KK489 pKa = 7.17TQKK492 pKa = 10.83DD493 pKa = 3.46YY494 pKa = 11.8EE495 pKa = 4.2EE496 pKa = 4.3RR497 pKa = 11.84KK498 pKa = 8.32RR499 pKa = 11.84TKK501 pKa = 10.71NMYY504 pKa = 10.08RR505 pKa = 11.84DD506 pKa = 3.31LKK508 pKa = 10.62LRR510 pKa = 11.84KK511 pKa = 9.11EE512 pKa = 4.2GLFAVV517 pKa = 4.53
Molecular weight: 60.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1577 |
114 |
586 |
394.3 |
44.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.531 ± 1.915 | 0.698 ± 0.283 |
6.468 ± 0.74 | 5.834 ± 1.023 |
5.263 ± 0.987 | 6.595 ± 0.678 |
1.458 ± 0.486 | 5.073 ± 0.396 |
7.229 ± 1.429 | 7.546 ± 0.553 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.029 ± 0.196 | 6.278 ± 0.805 |
3.044 ± 0.769 | 4.375 ± 1.445 |
4.629 ± 0.94 | 8.117 ± 1.114 |
6.785 ± 0.241 | 4.629 ± 0.331 |
0.888 ± 0.302 | 6.531 ± 1.46 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
